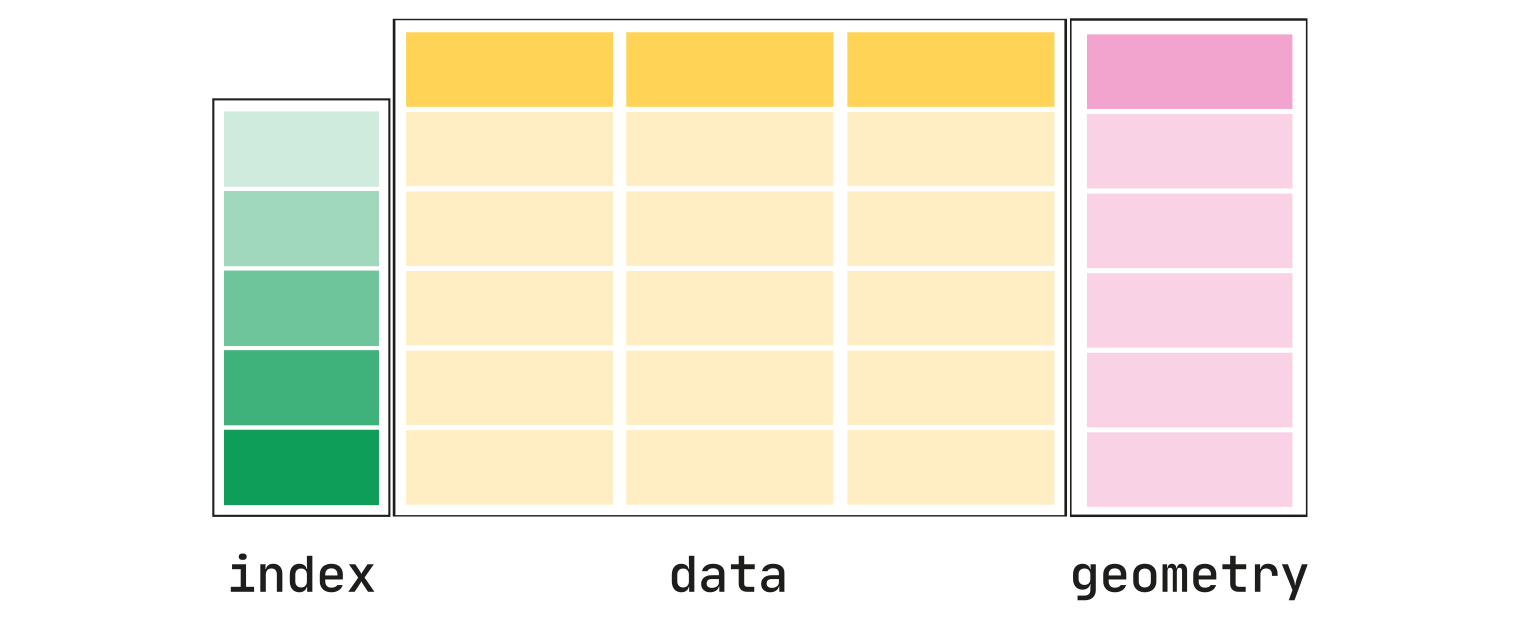
10. Geopandas#
Geopandas enables the use and manipulation of geospatial data in Python. It extends the common datatype used in pandas to allow for the many and unique geometric operations: GeoSeries and GeoDataFrame. Geopandas is also built on top of shapely for its geometric operation; its underlying datatype allows Geopandas to run blazingly fast and is appropriate for many machine learning pipelines that require large geospatial datasets.
Instalation#
Following steps:
1. create new environment#
conda creare -name new_env
2. Activate new environment#
conda activate new_env
3. Install package#
conda install -c conda-forge python=3 geopandas
Geopandas data types#
Shapefile (.shp): vector data that stores geospatial information such as topology, shape geometry, coordinate etc
GeoJSON (.geojson): geospatial information and relevant characteristics (object or string)
Finally, Geopandas creates an additional geometry column
Coordinate Reference Systems (crs)#
The coordinate reference systems tell us how GeodataFrame o GeoSeries relate to places on Earth
relevant operations: setting a projection and re-projecting#
setting projection: geopandas.read_file() most of time includes projection information. Check using “Geoseries.crs” If GeoDataframe does not include projection, hence use data[“geometry”].set_crs(epsg=4326) for WGS-84.
WGS-84:#
The Global Positioning System uses the World Geodetic System (WGS84) as its reference coordinate system. It’s made up of a reference ellipsoid, a standard coordinate system, altitude data, and a geoid
Reprojecting:#
In somes cases, due to distortions in bidimensional plane and your application of geospatial data, re-projection is required.
A commonly used default is Universal Transverse Mercator (UTM)
import pandas as pd
from pandas import Series, DataFrame
import numpy as np
import matplotlib.pyplot as plt
import chardet
import geopandas as gpd
from geopandas import GeoSeries
from shapely.geometry import Point, LineString
import folium
from folium import Marker, GeoJson
from folium.plugins import MarkerCluster, HeatMap
---------------------------------------------------------------------------
ModuleNotFoundError Traceback (most recent call last)
Cell In[1], line 5
3 import numpy as np
4 import matplotlib.pyplot as plt
----> 5 import chardet
6 import geopandas as gpd
7 from geopandas import GeoSeries
ModuleNotFoundError: No module named 'chardet'
# Gettting the character format (encoding type)
base = open(r'../_data/Datos_panel_distritos.csv', 'rb').read()
det = chardet.detect(base)
charenc = det['encoding']
charenc
'utf-8'
Covid Data#
# Import csv file from panel covid information at distric level
# Panel data from march-2020 to first two wees of january-2022
cv_data = pd.read_csv( r'../_data/Datos_panel_distritos.csv', encoding = charenc)
cv_data.head( 5 )
| Código | Departamento | Provincia | Distrito | Mes | Año | Casos | Muertes Covid | Muertes totales | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 10101 | AMAZONAS | CHACHAPOYAS | CHACHAPOYAS | 3 | 2020 | 0 | 0 | 16 |
| 1 | 10101 | AMAZONAS | CHACHAPOYAS | CHACHAPOYAS | 4 | 2020 | 47 | 0 | 5 |
| 2 | 10101 | AMAZONAS | CHACHAPOYAS | CHACHAPOYAS | 5 | 2020 | 165 | 3 | 11 |
| 3 | 10101 | AMAZONAS | CHACHAPOYAS | CHACHAPOYAS | 6 | 2020 | 119 | 2 | 15 |
| 4 | 10101 | AMAZONAS | CHACHAPOYAS | CHACHAPOYAS | 7 | 2020 | 175 | 2 | 8 |
cv_data.dtypes
Código int64
Departamento object
Provincia object
Distrito object
Mes int64
Año int64
Casos int64
Muertes Covid int64
Muertes totales int64
dtype: object
# rename variable to merge later
cv_data = cv_data.rename({'Código':'UBIGEO'}, axis =1 )
cv_data.head( 5 )
| UBIGEO | Departamento | Provincia | Distrito | Mes | Año | Casos | Muertes Covid | Muertes totales | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 10101 | AMAZONAS | CHACHAPOYAS | CHACHAPOYAS | 3 | 2020 | 0 | 0 | 16 |
| 1 | 10101 | AMAZONAS | CHACHAPOYAS | CHACHAPOYAS | 4 | 2020 | 47 | 0 | 5 |
| 2 | 10101 | AMAZONAS | CHACHAPOYAS | CHACHAPOYAS | 5 | 2020 | 165 | 3 | 11 |
| 3 | 10101 | AMAZONAS | CHACHAPOYAS | CHACHAPOYAS | 6 | 2020 | 119 | 2 | 15 |
| 4 | 10101 | AMAZONAS | CHACHAPOYAS | CHACHAPOYAS | 7 | 2020 | 175 | 2 | 8 |
Shape File#
# Upload shape file at district level
maps = gpd.read_file(r'../_data/shape_file/DISTRITOS.shp')
maps
| IDDPTO | DEPARTAMEN | IDPROV | PROVINCIA | IDDIST | DISTRITO | CAPITAL | CODCCPP | AREA | FUENTE | geometry | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 10 | HUANUCO | 1009 | PUERTO INCA | 100902 | CODO DEL POZUZO | CODO DEL POZUZO | 0001 | 1 | INEI | POLYGON ((-75.31797 -9.29529, -75.31710 -9.297... |
| 1 | 10 | HUANUCO | 1009 | PUERTO INCA | 100904 | TOURNAVISTA | TOURNAVISTA | 0001 | 1 | INEI | POLYGON ((-74.64136 -8.82302, -74.64036 -8.828... |
| 2 | 25 | UCAYALI | 2503 | PADRE ABAD | 250305 | ALEXANDER VON HUMBOLDT | ALEXANDER VON HUMBOLDT | 0001 | 1 | INEI | POLYGON ((-75.02253 -8.74193, -75.02267 -8.742... |
| 3 | 25 | UCAYALI | 2503 | PADRE ABAD | 250302 | IRAZOLA | SAN ALEJANDRO | 0001 | 1 | INEI | POLYGON ((-75.13864 -8.56712, -75.13956 -8.569... |
| 4 | 25 | UCAYALI | 2503 | PADRE ABAD | 250304 | NESHUYA | MONTE ALEGRE | 0001 | 1 | INEI | POLYGON ((-75.01589 -8.44637, -75.01585 -8.446... |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 1868 | 10 | HUANUCO | 1006 | LEONCIO PRADO | 100608 | CASTILLO GRANDE | CASTILLO GRANDE | 0001 | 1 | INEI | POLYGON ((-76.08083 -9.13017, -76.08026 -9.130... |
| 1869 | 10 | HUANUCO | 1006 | LEONCIO PRADO | 100609 | PUEBLO NUEVO | PUEBLO NUEVO | 0001 | 1 | INEI | POLYGON ((-75.88828 -9.00906, -75.88756 -9.010... |
| 1870 | 10 | HUANUCO | 1006 | LEONCIO PRADO | 100610 | SANTO DOMINGO DE ANDA | PACAE | 0001 | 1 | INEI | POLYGON ((-75.91141 -8.88593, -75.91182 -8.886... |
| 1871 | 21 | PUNO | 2111 | SAN ROMAN | 211105 | SAN MIGUEL | SAN MIGUEL | 0001 | 1 | INEI | POLYGON ((-70.13203 -15.33382, -70.12355 -15.3... |
| 1872 | 15 | LIMA | 1508 | HUAURA | 150806 | HUAURA | HUAURA | 0001 | 1 | INEI | POLYGON ((-77.37630 -10.89734, -77.37604 -10.8... |
1873 rows × 11 columns
# Plot 1 district (its a collection of points)
maps["geometry"][100]
## Set of points for first district
maps.geometry[0].boundary.coords.xy
(array('d', [-75.31797010604475, -75.31710488392423, -75.31604648791898, -75.31537303628215, -75.31469932241646, -75.31392881111611, -75.3133303495971, -75.31169723519622, -75.30615235974136, -75.30464848475174, -75.30204673484673, -75.30106448524998, -75.29776760924966, -75.29447260989667, -75.28983985902022, -75.28862698442924, -75.2863086102684, -75.2851563586247, -75.28393560955703, -75.28272260912615, -75.28220311024171, -75.28202736045313, -75.28138673398348, -75.28156248370055, -75.28098236019497, -75.28017386006405, -75.27959373487306, -75.27774798404425, -75.27780860999633, -75.27711523520276, -75.27648235912312, -75.27555073511627, -75.27393360884412, -75.27306448398218, -75.27278123480181, -75.27197261018847, -75.27063673497003, -75.26936310921559, -75.26861523478374, -75.26768548488297, -75.26531248414989, -75.26329098403181, -75.26230660998112, -75.26178710908653, -75.26045898389629, -75.2592461099199, -75.25808785916968, -75.25611910895401, -75.25409760880586, -75.25021485905677, -75.24755860988529, -75.24643748485443, -75.24575198510132, -75.24066798386781, -75.24050198407201, -75.2372597351648, -75.2359842337778, -75.23378711019097, -75.22974410985302, -75.226158234026, -75.22216785915634, -75.21956635905468, -75.2180019840473, -75.21586523510524, -75.21453123508553, -75.21216472468012, -75.20932810907095, -75.20695510989407, -75.20516211010215, -75.20215623501099, -75.2003104839743, -75.19660160978003, -75.19516011003475, -75.19290235888997, -75.19209373494306, -75.18868161016542, -75.1849726101953, -75.18167985885947, -75.17936723501833, -75.17792573421747, -75.17651361007904, -75.17580660879213, -75.17588473404857, -75.17943548397147, -75.18314448413241, -75.1839609850594, -75.18460160999712, -75.1869727339589, -75.19109373402421, -75.19451173489051, -75.19659360997343, -75.19792973499995, -75.20158398488337, -75.20500198504806, -75.20667973505584, -75.20683598379895, -75.2068359841013, -75.20659960977974, -75.20596873518195, -75.20596873483112, -75.20612698407149, -75.20633786011939, -75.20691598519164, -75.20854098398303, -75.20854098480078, -75.20779485889199, -75.20640623490311, -75.20553710915271, -75.20530660914335, -75.20536723514675, -75.20503323488936, -75.20426948392367, -75.20127735983812, -75.20000386022781, -75.19913473392899, -75.19899998492298, -75.1989999851395, -75.19905273482743, -75.19797460903715, -75.19750973410777, -75.19659360985185, -75.19443548410442, -75.19213085881688, -75.18875198418357, -75.18643948386506, -75.18351760983256, -75.18044335995273, -75.17852723516307, -75.17688860993508, -75.17600198381751, -75.17480660900958, -75.17352148391444, -75.173218733991, -75.17999223413648, -75.1822968599235, -75.18338285977404, -75.18724011009328, -75.18908786007069, -75.19039060880829, -75.19096286020743, -75.19353323494937, -75.19743548407143, -75.19924410995401, -75.20067385988443, -75.20143360996087, -75.20238673386291, -75.2050527347858, -75.20847848494634, -75.21028710979664, -75.21257223488152, -75.21552348416702, -75.21771285920863, -75.22187111011863, -75.23419135979907, -75.23463673377455, -75.23518748483019, -75.23652735908092, -75.23967573410738, -75.24243361003971, -75.2444022351147, -75.24629298498576, -75.24789448494153, -75.26144335922277, -75.27930473392661, -75.28793360883787, -75.28823823413148, -75.29363286020288, -75.2971503589057, -75.29998636014335, -75.30324998406797, -75.30473823498636, -75.30872261020085, -75.32629486011592, -75.32875786018633, -75.32968161005624, -75.33107810920109, -75.33108598486422, -75.3306269839544, -75.32939848519595, -75.32828710997478, -75.3277267338919, -75.32701760912431, -75.32457598521671, -75.32333786004773, -75.3144843599402, -75.31063861013075, -75.31059811012011, -75.31036961002894, -75.3099113588807, -75.30464685988397, -75.30306961015356, -75.3015653588925, -75.29987885993188, -75.29869360882827, -75.2978274851356, -75.2976104850456, -75.29702735980004, -75.29684485990714, -75.29620773493508, -75.29461510994275, -75.29413723521441, -75.29397798413402, -75.29406998505561, -75.29413710992816, -75.29371711011817, -75.29329523495892, -75.2926349851545, -75.2920911099713, -75.29140961002486, -75.29070073512855, -75.28952848482243, -75.28848048388858, -75.28622686015348, -75.28452811010743, -75.2826168590953, -75.28059948521934, -75.27921898499596, -75.27879435985193, -75.27898735995848, -75.27846048508178, -75.27804448519782, -75.277146358778, -75.2767089851593, -75.27582160978608, -75.27565148391722, -75.27573661009535, -75.27633823508313, -75.2770004851223, -75.27820148478185, -75.27988510990622, -75.28039498501522, -75.28053398414045, -75.28097485913963, -75.2818516099897, -75.2827113598492, -75.28308223483208, -75.28328010910735, -75.28330148502164, -75.28289686012033, -75.28255960912901, -75.2823742351136, -75.28240798491822, -75.28286736016344, -75.28372298409442, -75.28485248416462, -75.28581348385546, -75.28658823479486, -75.28711160889145, -75.28738135893104, -75.2878028592084, -75.28793773499223, -75.28822436008011, -75.28969285889417, -75.29427273463745, -75.29642098397909, -75.29816010999048, -75.30072711013804, -75.30264086029058, -75.30359773454632, -75.3048264854433, -75.30609173539113, -75.31165173520435, -75.31715398387632, -75.32122073492371, -75.32520785871282, -75.33031135891332, -75.33868123504931, -75.34591348387505, -75.3473947337634, -75.34827310920116, -75.35019173525853, -75.35361935862846, -75.35667610883564, -75.3597678587384, -75.36394936009009, -75.36759648536662, -75.36935598512645, -75.37178748460849, -75.37354711029434, -75.37617060984354, -75.37818598464753, -75.3797214846266, -75.38125235997052, -75.38326011006365, -75.38957060880614, -75.39575498515558, -75.39805323545329, -75.3995958597389, -75.40176185911275, -75.40320536031157, -75.40365185985263, -75.40590273418383, -75.4069273597745, -75.40712186031277, -75.40723660893207, -75.40948273466432, -75.41024723434549, -75.41076123363155, -75.4110596096462, -75.41118161010172, -75.41111385975114, -75.41108673527731, -75.41122223542993, -75.41131723530492, -75.41137148386235, -75.41134598514765, -75.41114548514474, -75.41038811043433, -75.40984285862646, -75.40932398543612, -75.40933748476363, -75.40951386008987, -75.40953823510546, -75.40990423523846, -75.4099639845656, -75.40960048527847, -75.40896423470821, -75.40791385968117, -75.40712735866333, -75.40609686016427, -75.40467311024338, -75.40394173478433, -75.40297298496995, -75.40177585993945, -75.39856086042477, -75.39769961035961, -75.39662335880439, -75.39635386016835, -75.39576223367652, -75.39554723500993, -75.39565460978434, -75.3961937337802, -75.3972152346812, -75.39787023360287, -75.39824048492227, -75.39854123542688, -75.39879573472376, -75.3988419853768, -75.39923360967936, -75.39974435994867, -75.40136398380565, -75.40232848494759, -75.4028128599154, -75.40361960993107, -75.40426210860898, -75.40443210973808, -75.40443773519479, -75.4044806099347, -75.40423823381838, -75.40441998486176, -75.40478361001448, -75.40584385895062, -75.40729785958649, -75.40975423457525, -75.41093786034173, -75.41212148493906, -75.41341248478682, -75.41443586031193, -75.41615848485645, -75.41820348468622, -75.41949573464126, -75.4199103604297, -75.42029485980646, -75.42078936017896, -75.42144210921509, -75.42223861002577, -75.42291635964813, -75.42445735920887, -75.42553923537434, -75.42630336029907, -75.42704398466981, -75.4281507341862, -75.42972786003861, -75.43080135978306, -75.43161586045062, -75.43226361041235, -75.43299948482411, -75.43355935998517, -75.43483648536227, -75.43579885976072, -75.43665048499793, -75.43732661018709, -75.43794611038135, -75.43844585896272, -75.43972048483874, -75.44076986011999, -75.44174061016753, -75.44270310966374, -75.44379523478216, -75.44466523504451, -75.44555048472455, -75.44699698505241, -75.44862436035669, -75.44955173543724, -75.44971836015242, -75.45006998544602, -75.45055123464925, -75.45076736031268, -75.45080273524076, -75.45089523478225, -75.45127373533575, -75.4517657346359, -75.45225773496037, -75.45237123475147, -75.45244698384823, -75.45309036032484, -75.45369598516152, -75.45502060999303, -75.4560803597418, -75.45713998537838, -75.45907023456624, -75.46088248539837, -75.46230860971542, -75.46210011020935, -75.46190010997071, -75.46243348483972, -75.46350011005666, -75.46436673490888, -75.46436673414615, -75.46470011026003, -75.46576673376478, -75.46623335882992, -75.46610010958585, -75.46590010995192, -75.4668333595601, -75.4680999848166, -75.46796673467662, -75.46769998513514, -75.46709998527325, -75.46777598518912, -75.46964536030356, -75.47151473492283, -75.47322823474343, -75.47455248474012, -75.47595448471213, -75.47657760973794, -75.47774598536569, -75.4782912347656, -75.47945948457635, -75.4803163601722, -75.48195198506824, -75.48249723497719, -75.484522485154, -75.48608023460395, -75.48740435991613, -75.48950748496452, -75.49075373462728, -75.49129898541742, -75.49207785957458, -75.49254523417494, -75.49371348524107, -75.49511560992966, -75.49620598510171, -75.49753023384773, -75.49885436016893, -75.50196998412343, -75.50469610893845, -75.50625398489291, -75.50801998544216, -75.50837360925715, -75.50831510879806, -75.50858448507341, -75.50897823503539, -75.5092199852275, -75.50972873506095, -75.51051173460081, -75.51148573491048, -75.51221111021556, -75.51351673429292, -75.51471861021423, -75.51554760959579, -75.51701898541563, -75.51894623387494, -75.52054198519073, -75.52155748493783, -75.52253148529621, -75.52381623530039, -75.52450010918596, -75.52908010985163, -75.53009548531764, -75.53111098482958, -75.53220935957046, -75.53297611039329, -75.53410123528619, -75.53555623534112, -75.53710223527628, -75.53864811002649, -75.54164911042699, -75.5439226088464, -75.54482585964507, -75.54601410985396, -75.54719636008059, -75.54837860986044, -75.54955598458685, -75.55058160982668, -75.5512422353138, -75.55216398496196, -75.55335610980427, -75.55460623458879, -75.55549460962976, -75.55700560972866, -75.55797873459807, -75.55892760973177, -75.55962098473145, -75.56014211024382, -75.56073973535649, -75.56020048530877, -75.56029148475864, -75.56038236002308, -75.56183748538336, -75.5656568603763, -75.56902161043342, -75.5720224848053, -75.57629660986146, -75.57911573532618, -75.58054211012188, -75.58197535966644, -75.58448110997813, -75.58648173539945, -75.58848235992087, -75.59039211010992, -75.5913923600753, -75.5922108600393, -75.59227336002749, -75.59291386001468, -75.59286398521874, -75.59257048488985, -75.59239373516067, -75.59233411011763, -75.59297360997436, -75.59343886003762, -75.59431198516182, -75.59530248420816, -75.59617673499974, -75.59681848481108, -75.5983934848523, -75.59955923505998, -75.60060823519802, -75.60183148513516, -75.6030547351861, -75.6042788600522, -75.60544485991232, -75.60631960986916, -75.60818448506814, -75.60975773515781, -75.61127386011128, -75.61261473492596, -75.61401298494218, -75.61552798503163, -75.61710035980906, -75.61826510986765, -75.61937198502488, -75.6204208602091, -75.6213522348794, -75.62240048516497, -75.62344798487959, -75.6243803599126, -75.62513836001483, -75.62624648508114, -75.62700548507519, -75.6275904848062, -75.62829185999777, -75.62911010987182, -75.62992785983994, -75.63097736011406, -75.63214335979967, -75.6344163600042, -75.63569873477462, -75.63709786015482, -75.63878673510033, -75.6398352348811, -75.641408609888, -75.64298286002634, -75.64395223521063, -75.64407985985417, -75.64508298511856, -75.6461331101425, -75.646833610018, -75.64770798507357, -75.64922348490478, -75.65097073518012, -75.65312573508305, -75.65580448496434, -75.65813373492864, -75.65941448491242, -75.6610442350484, -75.6625584850126, -75.66413148498935, -75.66453911004163, -75.66541086011705, -75.6671593600538, -75.66914098500166, -75.67106385979503, -75.67275398515989, -75.67327748494324, -75.67373923481051, -75.67397098521116, -75.67449373491723, -75.67577298479387, -75.67682035985518, -75.67780886004152, -75.67809723509993, -75.67867735987608, -75.67902485986934, -75.67890585986083, -75.67931211021875, -75.68012548512803, -75.68041536007422, -75.68035523508718, -75.68052873482878, -75.68140061010126, -75.68158273479429, -75.68165623504422, -75.68175198485199, -75.68186460983412, -75.68296948496072, -75.68448185992406, -75.68535436003754, -75.68710110987678, -75.68849773485096, -75.69082673522013, -75.69315573508464, -75.69368011008095, -75.69376536013476, -75.69422073520138, -75.69522260993631, -75.69564848506911, -75.69567848484218, -75.69571910989958, -75.69746735983449, -75.69898373510796, -75.70027148498258, -75.70044223480329, -75.70172623492897, -75.70312661016555, -75.70487523502021, -75.7060413601518, -75.70755598479155, -75.70930335996127, -75.71040786005534, -75.71157111012343, -75.71291073508706, -75.71448323492461, -75.71564736006462, -75.71686873514182, -75.71791598514778, -75.71966286001629, -75.72088448510175, -75.7223958601686, -75.72379223492669, -75.72483985999133, -75.7256546097928, -75.72681910990326, -75.72850498477266, -75.72955123519398, -75.73042273513494, -75.73088623503952, -75.7312343601107, -75.73204773513982, -75.73321085996656, -75.73478110998428, -75.73605998502512, -75.73681511004594, -75.73739573503872, -75.73803261003349, -75.73826135993556, -75.7386674848313, -75.73959673490145, -75.73967735983473, -75.74023598489208, -75.7412844849934, -75.74303273517154, -75.74443135999718, -75.74559710986154, -75.74746173477627, -75.74845298514634, -75.7496777349869, -75.75061011013554, -75.75258911001544, -75.75270923587118, -75.75305073505905, -75.75308011002426, -75.75299410982939, -75.75242386005834, -75.75228123509731, -75.75213861012418, -75.75196673493826, -75.75179486001304, -75.75170898492291, -75.75182423499086, -75.75222260994042, -75.752509735091, -75.75256635983811, -75.75267773502259, -75.75285736013006, -75.7528573599568, -75.75282036001323, -75.75282035987976, -75.75292773491458, -75.75307223511686, -75.75303511006462, -75.75332223502642, -75.75410935992568, -75.75478910989654, -75.75543361011682, -75.75572073495346, -75.75582810978818, -75.75582810985782, -75.75600798613056, -75.75661523514924, -75.75738673517895, -75.758089860121, -75.75875198495851, -75.75910160993931, -75.75937498516845, -75.75945310996974, -75.75949223505984, -75.75921873506474, -75.75917973499912, -75.75945311020342, -75.7599198598885, -75.76054486008678, -75.76089448599288, -75.7611542352114, -75.76140823486566, -75.76131248508227, -75.7613339852187, -75.76142973514942, -75.76137698620545, -75.76134373484342, -75.7612187348538, -75.76068161014393, -75.76022848513759, -75.75983785978156, -75.75940623498553, -75.75897473487925, -75.75862710982926, -75.75841598486596, -75.75814260987079, -75.75790035982152, -75.75728911015614, -75.75662498485383, -75.75633010995301, -75.75604485993773, -75.75574023498234, -75.75525585987799, -75.7548769849056, -75.7545391101711, -75.75435935990521, -75.75420110994321, -75.75422260980804, -75.75416985980979, -75.75430660978475, -75.75457035995385, -75.7551718600298, -75.7554667351273, -75.75550785998246, -75.75533011004859, -75.75487698496795, -75.75438086004368, -75.75410735991574, -75.75369536010538, -75.7534003600299, -75.75292773513058, -75.7525683600482, -75.75221098517244, -75.75197848498277, -75.75152535981783, -75.75125186000747, -75.75110548481416, -75.75115423490666, -75.75122848489104, -75.75158598490702, -75.75209173521029, -75.75240423522166, -75.75252348477261, -75.75258598480582, -75.75269335977652, -75.75279098496401, -75.75292573503282, -75.75293361019686, -75.75292186003566, -75.752882860157, -75.75279885982275, -75.75262886021494, -75.7524551101717, -75.75231048486546, -75.7521757348295, -75.75204685982465, -75.75191211021593, -75.75183786001583, -75.75186323482819, -75.75183010994044, -75.75169135980633, -75.75156835986718, -75.75126948605396, -75.75107610981752, -75.75083398482758, -75.75055860994166, -75.75040623502952, -75.7500683600964, -75.74966798505113, -75.7492363598297, -75.7489374848667, -75.74862698517353, -75.74815036002315, -75.74737685997462, -75.74678123495528, -75.74623236001315, -75.74552923514159, -75.74497660987595, -75.74480473512284, -75.74450585990468, -75.74420698517906, -75.7440369849212, -75.74397261016935, -75.74397260994323, -75.74356836010179, -75.7427578597975, -75.7416562350524, -75.74044535981926, -75.73954673503135, -75.73859173494577, -75.73759561007313, -75.73649798515049, -75.73557223490286, -75.7342773599637, -75.73254498498324, -75.73151173489975, -75.73009173506563, -75.72892773482266, -75.72779885983778, -75.72681635990676, -75.72597848484378, -75.72504686018385, -75.72466986011608, -75.72446873502525, -75.72419335992834, -75.72383986014546, -75.72284173501333, -75.72216986006299, -75.72120111014544, -75.7203516100397, -75.71956248507934, -75.71891985990607, -75.71795323515562, -75.7175547350019, -75.71701173504198, -75.7165507350652, -75.71628323515517, -75.71632611011529, -75.7165058597995, -75.71673048494057, -75.71704298521075, -75.71704298519461, -75.71690823505128, -75.71668548484485, -75.71659573509947, -75.71678911000964, -75.71718548479578, -75.71742973484663, -75.71749023487428, -75.71726561015271, -75.716744109979, -75.71637110992049, -75.71571673483237, -75.71511910977756, -75.71440023487253, -75.71411136013853, -75.71416410996022, -75.71450786007455, -75.71479098495763, -75.71408785986813, -75.71483598480512, -75.7151191102069, -75.71591210987003, -75.71642386001201, -75.71733785979575, -75.71889448518027, -75.72177923479029, -75.72299998483527, -75.72351348501473, -75.72423236010343, -75.72450386004095, -75.72454485986373, -75.72450385985826, -75.72435161008292, -75.72429485977312, -75.72444735997038, -75.72450198480703, -75.72431835982621, -75.72420710987768, -75.72376173496906, -75.72347261022759, -75.72323636014062, -75.72333598501922, -75.72298435981799, -75.72182423508491, -75.72100786018177, -75.72094711012204, -75.7212363600305, -75.72123048504477, -75.72094711016439, -75.72011898522715, -75.71958598478221, -75.71905273517064, -75.71834173481062, -75.71705073496327, -75.71658398512278, -75.71640823502378, -75.71373823500795, -75.71147073498497, -75.71151761009338, -75.71186135994219, -75.71454685997873, -75.71442385994618, -75.71343160999533, -75.71249410985348, -75.71231836008707, -75.71231048518274, -75.71294336000614, -75.71599611008139, -75.71656836010061, -75.71661323491213, -75.71614060999497, -75.7153847348842, -75.71508785999072, -75.7142538598282, -75.71360160987263, -75.71324610990939, -75.71330661016174, -75.71345510997764, -75.7134551101957, -75.7137382351965, -75.71373823499107, -75.71379098478695, -75.71388286009397, -75.71385160978957, -75.71309573516481, -75.7129766098426, -75.71308973504932, -75.71267773492798, -75.71255461021303, -75.71092973497764, -75.71028910991242, -75.70976948506546, -75.706292985083, -75.70460548512153, -75.7039101101887, -75.70315623481832, -75.70072261010563, -75.69997661002452, -75.69957223487853, -75.69900786010987, -75.69834173483525, -75.69769548485817, -75.69626173508044, -75.69521285993193, -75.69404098501298, -75.6930722349012, -75.69254885990256, -75.69125598513682, -75.69044923499848, -75.6886523601217, -75.68778510997902, -75.68701760981838, -75.6853711098887, -75.6813729849465, -75.68038861020636, -75.67957223516186, -75.67905473512968, -75.67811523513728, -75.67758985992674, -75.67769535978204, -75.6784354851406, -75.6781992349197, -75.67772660981319, -75.67691798487249, -75.67527735992383, -75.67347661008301, -75.67243161003898, -75.67201948514844, -75.67172260978555, -75.67061523516014, -75.66924223487547, -75.66895698501804, -75.66874798497321, -75.66819135997531, -75.66758598494192, -75.66539848479488, -75.6637109852195, -75.66307811000041, -75.66220311020146, -75.66125586010446, -75.66093948521444, -75.66093948515669, -75.66102536002296, -75.66157035987965, -75.66183010988732, -75.66188660988576, -75.66162886007936, -75.66105473501108, -75.6608242351088, -75.66079685998398, -75.66030873515308, -75.65956248491489, -75.65818361006404, -75.65740236013824, -75.65583786007761, -75.65478510981154, -75.65350385990553, -75.65268748486312, -75.65141211020676, -75.64908598497408, -75.64414261009733, -75.64106823498446, -75.63636135988133, -75.63363085986379, -75.63316728026996, -75.63252348493648, -75.63135348514993, -75.62989848498404, -75.62977535991757, -75.63005861008236, -75.63005860979604, -75.62871485980679, -75.62777736009878, -75.62771673499572, -75.62789061022647, -75.62863861016302, -75.62915023489398, -75.62917748675841, -75.62916997958547, -75.62836129960783, -75.62719111614729, -75.62621517663877, -75.62514202688055, -75.62407131064197, -75.62309606303006, -75.62192500162826, -75.62065724753819, -75.61919488748379, -75.61743967126426, -75.61665987815371, -75.6154883002344, -75.61392614377598, -75.61236331093392, -75.61255587126811, -75.6122584216995, -75.61137767406547, -75.60942668413679, -75.60757460347884, -75.60669911198505, -75.60543185970874, -75.6045546702959, -75.60367899193693, -75.60338883389153, -75.60270692172327, -75.60065949925917, -75.59948794103809, -75.59812026577288, -75.59665370982185, -75.5961639214654, -75.59440554132641, -75.59372213410245, -75.5917698069918, -75.58952497585693, -75.58757331665487, -75.58630538412558, -75.58454803101341, -75.5834726168618, -75.58220209648312, -75.5796646324709, -75.57810387444488, -75.57644603836741, -75.57449488657898, -75.57225042346272, -75.57039603602226, -75.56805402380633, -75.56514154843018, -75.56247326828276, -75.56042347056439, -75.55729962252144, -75.55437090860153, -75.55212557065249, -75.5505634431438, -75.54880697236837, -75.54695310053563, -75.54587952704168, -75.54412230740951, -75.5413896881991, -75.53982834472862, -75.53826685478566, -75.53670342915865, -75.5351409103443, -75.53328410749606, -75.5308439213286, -75.5284043334625, -75.5259644583893, -75.52401105257468, -75.52166730549328, -75.51795856208633, -75.51542261746084, -75.51356953911744, -75.51161890370327, -75.50966798147311, -75.50781306054301, -75.50595927293477, -75.50293219897901, -75.49867973366031, -75.49682623514423, -75.49439060904342, -75.493644484833, -75.49260161017827, -75.48987498408724, -75.48912110986038, -75.48779298361984, -75.48668748506287, -75.48523635976835, -75.48407811034718, -75.48274223531996, -75.48256635880074, -75.48227536002537, -75.48140623474632, -75.47653910979011, -75.47329098493668, -75.47283198471678, -75.47132035914603, -75.47050198512369, -75.46975785883501, -75.46766598456958, -75.46674410996803, -75.46547073398978, -75.46505660859907, -75.46436323457431, -75.4607167339002, -75.4591444851759, -75.45827548528536, -75.45722111043534, -75.45625460868604, -75.45608198483109, -75.45605073520306, -75.45618360969269, -75.4558944847775, -75.45531448412197, -75.45513861028758, -75.45426173519832, -75.45403323479502, -75.45321686035061, -75.45298048516122, -75.4532089853156, -75.45361323464658, -75.45326761038524, -75.45193560956176, -75.451644484361, -75.45164448519154, -75.45199023412874, -75.45203679608483, -75.45196794360777, -75.45100224144926, -75.45037906889866, -75.44956407114836, -75.44913302667055, -75.44788673971044, -75.44716762282849, -75.44654499175869, -75.44587405606467, -75.44525094046287, -75.4446755252809, -75.44376452177306, -75.44256557309342, -75.44189439638569, -75.44107987301021, -75.4404567557794, -75.43968957654192, -75.43897004219679, -75.4382984503373, -75.43786626947468, -75.43690685677416, -75.43609156422639, -75.43426998131042, -75.43297580688039, -75.43144175028146, -75.43029116373259, -75.42971604463162, -75.4289493343299, -75.42808674027373, -75.42731910310255, -75.42664775381189, -75.42578469802845, -75.42482524359707, -75.42386573464819, -75.42285834494237, -75.4219948377712, -75.42103539433475, -75.41964460031926, -75.4184940219069, -75.41753548850772, -75.41672100147451, -75.41624238017948, -75.4156671447322, -75.41475621007783, -75.41403782278826, -75.41322360649673, -75.41236116716203, -75.41130696050317, -75.41058822962702, -75.40958162754356, -75.4089112222727, -75.40828875601522, -75.40781011234998, -75.40733146689837, -75.4070445319552, -75.40685347986556, -75.40627850654647, -75.40565553540455, -75.40474518490339, -75.40367045512045, -75.4022527413498, -75.40129368135962, -75.4004780694616, -75.39975866828689, -75.39884787760224, -75.39831986868165, -75.39745626819916, -75.39611352000003, -75.39443582137369, -75.39314170885461, -75.39232696093359, -75.39060205285995, -75.38921283670982, -75.38715346627049, -75.38581217112176, -75.38475831697508, -75.38322544360176, -75.38173983122198, -75.38049346604112, -75.37895872294717, -75.37728092115395, -75.37665837503317, -75.37617955446751, -75.37488574669555, -75.37339995834861, -75.37215373229958, -75.37095464405996, -75.3700431186806, -75.36874780075868, -75.36750053125041, -75.36601320484408, -75.3645741259848, -75.36327981546873, -75.36222570855426, -75.36155504471174, -75.3608358008733, -75.35968502115674, -75.3586789936096, -75.35781616190474, -75.35676097717288, -75.35575354541896, -75.35469870212944, -75.35340318767044, -75.35244385249575, -75.3515326053128, -75.35055112946156, -75.34949079824702, -75.34891202176135, -75.34842914224646, -75.3488129043846, -75.3484264900456, -75.34717325261175, -75.34601600812852, -75.34505197095433, -75.34428066690452, -75.34485755103026, -75.34456742476965, -75.34302557807132, -75.3416762762335, -75.34071206750946, -75.34061468408953, -75.34003594052803, -75.33916828883844, -75.33781817443219, -75.33646706216717, -75.3356960604757, -75.335020785783, -75.3340569675077, -75.33290105670028, -75.33097441072869, -75.32769883666481, -75.32480879239716, -75.32201492104804, -75.32066607654247, -75.31902865357705, -75.31797010604475]),
array('d', [-9.295288689326071, -9.297586845172532, -9.299406547690069, -9.30065156092951, -9.301609400474774, -9.302088700656896, -9.301537673150904, -9.30261722686015, -9.304294977423012, -9.3050389772132, -9.306884727071406, -9.307402353148234, -9.308607477017233, -9.31028322673086, -9.312123101588156, -9.31224222829104, -9.311890601627624, -9.31137697756975, -9.310464727127624, -9.309831976742544, -9.309199226549595, -9.30805072706903, -9.306730477602457, -9.305462851959419, -9.304486353125014, -9.306097602971477, -9.306613226815784, -9.307013727898493, -9.309773476985868, -9.311156227203924, -9.3137988531509, -9.314947227828043, -9.316097601816978, -9.317193352102095, -9.317880851892426, -9.318513728192386, -9.318859353046093, -9.31885935222249, -9.318400351860532, -9.318290977847653, -9.318980477115929, -9.319037102847233, -9.319732477110279, -9.320593602826591, -9.32191410297537, -9.322665977777206, -9.323064477110563, -9.323238353090801, -9.323068351984205, -9.32180472702862, -9.322095727212798, -9.3224257278207, -9.322665976876578, -9.32490822684042, -9.32497660208685, -9.326585976948309, -9.327040976956653, -9.328373103134311, -9.330265603041028, -9.332628852789924, -9.33578710306108, -9.338494102147024, -9.34039447807362, -9.343779351870865, -9.345453102992735, -9.348120763753286, -9.351318352969884, -9.353615227935677, -9.355164101898026, -9.357353477922913, -9.358449226840605, -9.35988085316393, -9.36063285302572, -9.362244102178636, -9.36264260200005, -9.365402351796376, -9.367593727156589, -9.370869102850065, -9.372824227114108, -9.374380852140575, -9.37560747787552, -9.376789101980819, -9.378679726881103, -9.395634602793507, -9.407708977009406, -9.41195897717057, -9.417476602854833, -9.425632851834393, -9.43729885294539, -9.444703102996982, -9.449939476891984, -9.452751976780803, -9.462056601901338, -9.469816351872169, -9.472798853194263, -9.4735801021763, -9.474447226986229, -9.475548852223028, -9.476652353101258, -9.477517601913064, -9.479093603059539, -9.480732477953609, -9.484005853137944, -9.488716852918799, -9.490154352830473, -9.49193747693954, -9.494066352219079, -9.496189477146402, -9.497519477194828, -9.500042977164242, -9.504339852076043, -9.507087853184544, -9.511201227912068, -9.51367772716884, -9.515687478194593, -9.517427727098607, -9.519236352793314, -9.522513727971045, -9.525568352124537, -9.529232478087692, -9.53121872698398, -9.533203103186622, -9.534121102877293, -9.534581977019968, -9.535652352209524, -9.538095728053577, -9.538861351936873, -9.53995697702453, -9.54150785193417, -9.54252535290209, -9.543632852029532, -9.544917978033538, -9.546195353148619, -9.552755852196299, -9.556419977064422, -9.559931602092377, -9.585427726952616, -9.592603477820774, -9.595681601891613, -9.596632852149924, -9.59891797795592, -9.601583977994665, -9.603677728073478, -9.606154352809085, -9.607771477000824, -9.60881835287222, -9.609960851897313, -9.610722602783028, -9.611675727786405, -9.612722601874566, -9.615005853111143, -9.615863228100466, -9.618847603121253, -9.62860935316263, -9.629753852047486, -9.630777353220083, -9.632273478031323, -9.635818352213846, -9.638574227151992, -9.64054297795525, -9.641802726840979, -9.643564477148061, -9.65408985314258, -9.67239847695987, -9.680177728062631, -9.680331976838204, -9.685670103143138, -9.68879297817017, -9.691488228167486, -9.695742227981839, -9.697728478013858, -9.700316226779421, -9.723353478217518, -9.72732422697652, -9.729611353038194, -9.736175727957288, -9.743962852940342, -9.747781226975349, -9.751753852985303, -9.753955103072656, -9.755132852098257, -9.756314477810292, -9.75977935288995, -9.760902351974039, -9.771003853056698, -9.774671853028451, -9.775015602840142, -9.775525226833395, -9.776104601887482, -9.781847103100745, -9.78362210278425, -9.785810351830207, -9.788590852832215, -9.791371476929953, -9.793604978058227, -9.794262102780692, -9.797296978111206, -9.799776478050433, -9.801846977963237, -9.805297727940626, -9.80641260211263, -9.807899102208353, -9.81038022791654, -9.812022976982007, -9.814186976887129, -9.81508935282369, -9.816171477773707, -9.8169099778072, -9.817958353193433, -9.81870897807363, -9.820349977153967, -9.82174747685076, -9.824861226840767, -9.827568728137521, -9.83048872821729, -9.834788978097833, -9.838239727906512, -9.841265853078369, -9.843581228089022, -9.84559097797711, -9.84651985289446, -9.847821977802084, -9.848656227978779, -9.84976410315278, -9.85063310296755, -9.85206985314045, -9.854354978030528, -9.855919351885518, -9.857122477850453, -9.858444852067407, -9.859055603158424, -9.859197851837223, -9.85964835289974, -9.860390102905512, -9.860980228167932, -9.861452227036295, -9.861999476788299, -9.862531352935434, -9.863020228035452, -9.863348977920793, -9.864006477931412, -9.864731352784645, -9.865330728190543, -9.865742977967102, -9.866063228044027, -9.866316103195516, -9.866483853153113, -9.867024227074523, -9.86764810301213, -9.869418227947563, -9.870278103201201, -9.870699603160062, -9.8707958525589, -9.871095852667509, -9.870609852650034, -9.871350478259671, -9.87244385311385, -9.87324135283427, -9.873241353103026, -9.871787227781908, -9.871870101873014, -9.873958977956079, -9.8761119778776, -9.876909477778497, -9.876909477786853, -9.87555385167866, -9.874005227078896, -9.874478977680326, -9.874576102736057, -9.874575853435033, -9.87422410280351, -9.873403727666414, -9.872548353371826, -9.871802227810463, -9.871216853222393, -9.870960728039279, -9.870832602448615, -9.871152727750996, -9.871888478185326, -9.872240477222704, -9.872336352973264, -9.872432353411057, -9.872618603033288, -9.872921353266236, -9.87428860284433, -9.876528228039604, -9.877679103240894, -9.878455603320731, -9.87992222786994, -9.881285977619768, -9.881774102088343, -9.884651852727927, -9.885858102604061, -9.886003353192791, -9.886082602655506, -9.887151853115427, -9.887328477394192, -9.887585477797472, -9.888005728444796, -9.88842610302828, -9.888900728124213, -9.889931228379226, -9.890758351601676, -9.891666853392792, -9.892466853002816, -9.893032727893797, -9.893918977161093, -9.894645977703703, -9.894918601849781, -9.895504102561176, -9.89629060265787, -9.897076977609231, -9.897433228066632, -9.899164102749864, -9.90061410280589, -9.90200760295511, -9.902825602670362, -9.903178727858858, -9.90328722800291, -9.903422727673785, -9.903829602693747, -9.904059977609336, -9.9044897277617, -9.905267102802213, -9.90696435257911, -9.907772977635545, -9.908956478040523, -9.909548351863805, -9.911485853109014, -9.912776853437151, -9.913960477365663, -9.914876477563041, -9.916220227828319, -9.916801728432063, -9.917426476959918, -9.918120603137424, -9.918907227779812, -9.918953478369303, -9.919555853219322, -9.920341727793812, -9.921706853079058, -9.9226244769618, -9.923862728340907, -9.925098978232711, -9.926030728050819, -9.92651385307488, -9.92664672843199, -9.927546228241056, -9.928515602782568, -9.929424477714235, -9.930212103389579, -9.930636227842683, -9.930696852827339, -9.931073728044225, -9.9316108530424, -9.932848978082063, -9.936024853352706, -9.93785297811877, -9.939575602551997, -9.941298352780166, -9.942275852910257, -9.942634477018544, -9.94296685273622, -9.94325160310808, -9.943435103401157, -9.943558103232192, -9.943565977998162, -9.943106853218922, -9.942360852879347, -9.94206985295051, -9.941958602850264, -9.941880352603619, -9.94144047758499, -9.941014728040727, -9.94070010263647, -9.940366977872316, -9.93965335285651, -9.93920085336913, -9.938460353359801, -9.937942103050034, -9.937682977936845, -9.937538103085785, -9.93738685322927, -9.93720172785379, -9.936674478198995, -9.936366728412906, -9.936257728423584, -9.936442852587959, -9.936850103308018, -9.937257228420322, -9.937889478195416, -9.939228853284305, -9.940931977841615, -9.941551603435187, -9.942310477574381, -9.943032352965133, -9.943576228112533, -9.94373247832715, -9.943757853412933, -9.94505735333081, -9.946533477816956, -9.948387977332402, -9.949977603296418, -9.951756352749499, -9.95349735312675, -9.954897728302512, -9.955768227760245, -9.956222352792508, -9.956222352893917, -9.956903602763077, -9.957584852709374, -9.956803353142902, -9.957284477807207, -9.959169603034752, -9.960836352991066, -9.961636228046643, -9.963369603253113, -9.965969477827684, -9.967169477963466, -9.96950285314881, -9.971102728302867, -9.973569352664867, -9.975902727977939, -9.977835977676932, -9.979569352798478, -9.981169227901034, -9.982635853000772, -9.985502478349986, -9.9883691020014, -9.989058977825607, -9.989837852925234, -9.990694727722143, -9.991940977830382, -9.992174602625369, -9.990772603315406, -9.989526352637824, -9.988046353387972, -9.986722228037479, -9.985242352847422, -9.98508660310057, -9.983918228351342, -9.983061353335415, -9.982048853255549, -9.982048853402262, -9.982048853220192, -9.981815102661301, -9.981425728310853, -9.980724727966503, -9.980023602859092, -9.97994572822222, -9.979945728246832, -9.980023602713857, -9.980101603325465, -9.980101602907268, -9.979789978309498, -9.979166852613048, -9.979088978123343, -9.9790889776462, -9.979139978156695, -9.978627602678387, -9.977662477783213, -9.976916478248768, -9.977372352733767, -9.977969977735995, -9.978488228344077, -9.978843727769345, -9.979154602945528, -9.979506852858776, -9.980107852747551, -9.980605228391028, -9.981102602797193, -9.981371977416416, -9.981662102737346, -9.981599977760254, -9.981496353016269, -9.981206227704774, -9.9809161027448, -9.98064672812211, -9.981061227827809, -9.980957602813419, -9.980646728114209, -9.98070885265821, -9.980868352815719, -9.980868353360904, -9.98032272769899, -9.979958977651847, -9.980140853296916, -9.981050227676576, -9.981868727928429, -9.983349228135296, -9.984324103313497, -9.984687853143953, -9.98441497805527, -9.982998353363143, -9.982314602768781, -9.981693477865463, -9.980135727597485, -9.978286603187712, -9.976752478044773, -9.976024228243148, -9.975196853225668, -9.974418227987542, -9.973153102659467, -9.972629353395178, -9.97187222828802, -9.970112727728178, -9.9676823525798, -9.96604547770039, -9.964135728096267, -9.96322635284605, -9.962498852649503, -9.961316602561453, -9.960770977755146, -9.958952228073542, -9.959134102772396, -9.95941610270512, -9.958523977672428, -9.958133853423192, -9.9573153531436, -9.956405976672443, -9.955587478346445, -9.95467810339943, -9.952768477954976, -9.951962353126419, -9.950324602927651, -9.949603103203685, -9.948447978083152, -9.947408103157784, -9.946599227130037, -9.945731353171027, -9.945152727972982, -9.944515478065625, -9.944340353046698, -9.94433885289925, -9.944684352837669, -9.945374978173447, -9.945372978190164, -9.945313228111331, -9.944906602985668, -9.94455760280357, -9.944613227958598, -9.944784478068527, -9.94501397800775, -9.9448951030015, -9.944661102974408, -9.945062853038621, -9.945233727871312, -9.944884477909113, -9.944708353116596, -9.944011977839239, -9.943489728209727, -9.943198852919465, -9.943081352840068, -9.942443978111953, -9.941979727906471, -9.941168853171177, -9.941051477845008, -9.941223478054894, -9.94162585315189, -9.942260102916862, -9.94341460279702, -9.94445347806709, -9.945607603223777, -9.94653060297702, -9.946759727941185, -9.946873102965924, -9.946868851979445, -9.94692422679492, -9.94703710309733, -9.946340478176694, -9.945991727022601, -9.945873103169902, -9.946159102984836, -9.946561478208196, -9.946506353189932, -9.947079603055215, -9.947597602865642, -9.94817410193042, -9.948230227865508, -9.948227228193616, -9.947588227893903, -9.946832853167336, -9.945614103206026, -9.94456947808851, -9.94381572815837, -9.942714602222622, -9.94213372799868, -9.941841727966164, -9.941667601910012, -9.940394603148604, -9.94039110317948, -9.940387227938004, -9.940152228092927, -9.940148853097014, -9.939627728132848, -9.937430978190093, -9.936737102896075, -9.935869353138798, -9.934422102859855, -9.933610977790636, -9.932453353111214, -9.931008227791551, -9.929677977858148, -9.92863722792481, -9.927423977878505, -9.926614102841427, -9.925399103217671, -9.924647228170386, -9.92372285320323, -9.923086852954782, -9.921987228045055, -9.921515103202182, -9.92144535306221, -9.921339227906628, -9.920888352997679, -9.919730477884002, -9.918282852897562, -9.917471978221784, -9.91671722800561, -9.915732103180062, -9.914687228026253, -9.913700103142844, -9.913641227946925, -9.913688227934813, -9.913494227894262, -9.913212853001085, -9.91325397806969, -9.91326485197709, -9.913232477996246, -9.913228853154123, -9.913745727962407, -9.91440635282379, -9.914493977985671, -9.915357978127531, -9.916163978043471, -9.916275978073998, -9.916504602792088, -9.916270226795746, -9.915804352939915, -9.91447297807838, -9.91343035321023, -9.913080853015856, -9.912615227803258, -9.911919353085295, -9.910818852980583, -9.910007727914262, -9.90931047811415, -9.908325602942114, -9.906588852775144, -9.905545727901028, -9.904907853186387, -9.904386103047386, -9.903921227947535, -9.902068602847748, -9.900852852993182, -9.899637477795693, -9.898423102789195, -9.897728853059562, -9.896629228138067, -9.895528851948256, -9.894138602935072, -9.89269122797208, -9.891591727969228, -9.890665852844052, -9.888815478168965, -9.886850353089244, -9.886040478220195, -9.884651602820554, -9.884556602824375, -9.883898978221737, -9.883723353149605, -9.883719477938712, -9.883774227970093, -9.883887228011758, -9.883883103193511, -9.884111977970292, -9.884571478038708, -9.884569476957552, -9.883582727785551, -9.883516852864489, -9.883101601838746, -9.882644478095187, -9.881873102089287, -9.881130977826047, -9.880789103109862, -9.879818352963468, -9.87847660290754, -9.877333978174818, -9.87676372781948, -9.876220727847738, -9.875964852856498, -9.875621102977334, -9.875136727972347, -9.874236228135771, -9.87366410303548, -9.872734352886823, -9.87176760313423, -9.87080072781697, -9.87019135289421, -9.86954685182559, -9.868794978117762, -9.868509726918717, -9.868257853132661, -9.867757852959471, -9.867113227952302, -9.866826228156413, -9.86571685284133, -9.864070353225998, -9.863568351985407, -9.863390603202106, -9.863158227960469, -9.863001977850729, -9.862845727929539, -9.862650351898651, -9.862144476795663, -9.861404352827492, -9.860779353024327, -9.860273476989448, -9.859882853001807, -9.859611353066953, -9.859220728107005, -9.858910102937386, -9.858402352890096, -9.857873103197562, -9.85644335283979, -9.855705102992772, -9.855158228004825, -9.854345728104477, -9.853787102812134, -9.852923852839126, -9.852271478040677, -9.851787228221212, -9.851470727800859, -9.85126947679015, -9.850912101981748, -9.850291103002121, -9.849406227136058, -9.84887885302669, -9.848183603202756, -9.847675726846035, -9.846833978205837, -9.845949227815481, -9.845160227944689, -9.844265603086058, -9.843642602927105, -9.842937478105114, -9.842515478150403, -9.8420624779327, -9.841451228099118, -9.840818353207283, -9.839849603150128, -9.839249977092152, -9.838005852985646, -9.837036978119158, -9.835857477968545, -9.834855477835731, -9.83399222796885, -9.833675726958916, -9.83331835312913, -9.832980477819092, -9.832517602878568, -9.831105477804442, -9.830484353053635, -9.829968726804111, -9.829177727809054, -9.828535102919886, -9.827996103051817, -9.827007852974532, -9.82618547790372, -9.82485935286951, -9.824052728219385, -9.822876977157534, -9.821703102886943, -9.8207069781609, -9.819292977123553, -9.818898478150174, -9.818421852825168, -9.81788472814469, -9.817537103091581, -9.816982477809518, -9.816576228190513, -9.816113227876542, -9.815685602203416, -9.8155253531408, -9.815417977936931, -9.815337852919754, -9.815232478203086, -9.815050727829657, -9.814863228072397, -9.81388085295589, -9.813048851801796, -9.812869103074945, -9.812716728207818, -9.81259185189657, -9.812544977801565, -9.81255072815162, -9.812558602869387, -9.812585977986771, -9.812523478180822, -9.812406228124928, -9.812183603220092, -9.811933603006707, -9.811828102949107, -9.811546852913409, -9.811130852779968, -9.810580102033327, -9.809908227850542, -9.809523477780374, -9.808691353020606, -9.808265601994606, -9.807796852994898, -9.807603478156693, -9.80745510200802, -9.80741210293786, -9.8072422279904, -9.806773478065395, -9.806261726913874, -9.805706977998625, -9.805300726833934, -9.804728477825392, -9.804144478095509, -9.803730477787212, -9.80336135301468, -9.803175727826215, -9.802804727190846, -9.80240622795867, -9.802091853136346, -9.801923852880918, -9.801908227167758, -9.802343728214169, -9.803435601959052, -9.804417977917637, -9.805474728185061, -9.806529352826146, -9.807328103078873, -9.80742772807912, -9.807402351880626, -9.807249977993706, -9.806896477829639, -9.806031226884272, -9.805718728111628, -9.805374977857468, -9.804853478008852, -9.804347602790926, -9.804123102937751, -9.803974602809035, -9.803673851824442, -9.803289102884037, -9.802751978054346, -9.802304727949661, -9.80184185283008, -9.800783228025011, -9.800039102845517, -9.799337852910128, -9.798800728064577, -9.798205227849053, -9.7974746028965, -9.796937477773895, -9.796296852850674, -9.795435602820135, -9.794687477914863, -9.793941352917955, -9.793197228182976, -9.792585978115255, -9.79224222802327, -9.79191410180556, -9.791646478099665, -9.79128122800256, -9.790476602967203, -9.788070352920386, -9.786513728161083, -9.782208976894253, -9.778710977091805, -9.77699022819877, -9.77566797808663, -9.765673852851819, -9.762517602910862, -9.75849997696713, -9.753730478197042, -9.74878510190183, -9.746351603117034, -9.744412102052024, -9.741154353078969, -9.7404453529673, -9.739999977905768, -9.739763727105128, -9.73924810203414, -9.73875972814011, -9.738273478066048, -9.737601603224846, -9.735734353006762, -9.733427602160418, -9.731273477085836, -9.730124977073046, -9.728290977950508, -9.72076372820851, -9.719164102834952, -9.716290977084043, -9.71365235303631, -9.712738227138857, -9.711869102818014, -9.711009727917087, -9.70947660306598, -9.70805472795962, -9.707521477855721, -9.70604097803141, -9.704974602931543, -9.703306602989812, -9.702464852027447, -9.701839853205854, -9.697767603031878, -9.690771476934515, -9.688068352774621, -9.684621102042911, -9.66849022693981, -9.664121102793217, -9.660859352194564, -9.656437477920178, -9.655173851844722, -9.652531227888346, -9.650124978157805, -9.641499977903777, -9.6387481028205, -9.633349602021667, -9.629216852999377, -9.626234353120935, -9.624335977829674, -9.622566352036813, -9.62138085289115, -9.620611352816297, -9.619367353106554, -9.618714853076028, -9.616994102224965, -9.615447227023878, -9.613033226887296, -9.612572227070132, -9.61250010309152, -9.61246285189543, -9.610564478113872, -9.609824227909428, -9.607011728126338, -9.605628851797178, -9.603675728093322, -9.599546853180755, -9.597419977834079, -9.596626976929656, -9.593015602139417, -9.591011727990804, -9.589972477063043, -9.588253852981438, -9.584353476965589, -9.58256847784429, -9.581558603015992, -9.580408227112628, -9.579621102017143, -9.579136726797742, -9.578792977080354, -9.579056601934383, -9.579197226790598, -9.579076227012024, -9.578894476995913, -9.578550728072287, -9.578410103190436, -9.57824810313764, -9.577845727062709, -9.577380853011732, -9.575033102889847, -9.571080103168526, -9.569759728089139, -9.56815235312871, -9.567003853100914, -9.563849602095445, -9.560060602186525, -9.554951227067534, -9.547199228192085, -9.54421485286798, -9.541972601875273, -9.539451227009863, -9.530962853140977, -9.526316228207179, -9.522927727154524, -9.520748101921697, -9.517302727797414, -9.514374976876582, -9.511386727103513, -9.510037103150296, -9.509593726840933, -9.508826227926136, -9.507943353072108, -9.504915976877783, -9.502050603118164, -9.501527352895096, -9.501058602908074, -9.500628852074158, -9.500369102051934, -9.499738228181627, -9.499078103104807, -9.498130853180214, -9.497414103044946, -9.496695353132566, -9.496206978193698, -9.495173852841674, -9.494427727083405, -9.493539101807357, -9.493165978160391, -9.492906227944388, -9.492390601878355, -9.492005851840304, -9.490281227806747, -9.487408228177287, -9.48274410213304, -9.48056447713077, -9.47815235201, -9.474013728086504, -9.466597601887486, -9.461601601786299, -9.455107476900196, -9.450165978154644, -9.449106324857723, -9.44763472681268, -9.443665976853708, -9.437111352143745, -9.434697228121365, -9.433259727786776, -9.430781227775697, -9.42617960202682, -9.421234352995498, -9.41928122690058, -9.418357476916583, -9.416574228086063, -9.410939476844192, -9.410856810534028, -9.410856823911331, -9.41085826493088, -9.411150980376135, -9.410958958050799, -9.410960860958824, -9.412319043262528, -9.412514521938533, -9.412322833317717, -9.412615698802028, -9.41319953450386, -9.413686999150741, -9.414075874347759, -9.413593532175867, -9.412918109691077, -9.41185516956449, -9.410401668297936, -9.40768960535155, -9.406141086187802, -9.406241336821665, -9.407116430402425, -9.408571105901444, -9.40915454962169, -9.409640442778596, -9.41099823380597, -9.412451900314124, -9.413034333677786, -9.413812849287638, -9.413330452507864, -9.412267115048621, -9.410428914660542, -9.409267205447463, -9.407913877107546, -9.407624392916205, -9.406949523919877, -9.406372010744974, -9.406084629770476, -9.406280494037333, -9.405508382193716, -9.404153863988597, -9.402799663616975, -9.402222567368085, -9.40232200434076, -9.40271222828477, -9.402715407725736, -9.402331537416444, -9.401850143927447, -9.401466404770986, -9.401008199026478, -9.401184700238503, -9.400509807209744, -9.39925532522966, -9.398000508153018, -9.397035235042779, -9.396262640673683, -9.395974736047492, -9.395783852535033, -9.39549487552506, -9.39472255272051, -9.394048595365803, -9.393760346130819, -9.393375211580064, -9.391730643006667, -9.390667340166033, -9.38853880288525, -9.387670560784613, -9.387189816779369, -9.386515298567247, -9.38496812765925, -9.383324640881085, -9.382167534569874, -9.38246187641198, -9.38275520903926, -9.383048673179076, -9.383148367390277, -9.382182237799213, -9.381991133981955, -9.379960975302595, -9.37860547655157, -9.377812478057592, -9.377355351830658, -9.377011727339948, -9.376380852964871, -9.37402535199061, -9.373679727383376, -9.37265035242666, -9.371331977322711, -9.368683603402985, -9.367021477724734, -9.363062478415088, -9.361732477294652, -9.361505852660684, -9.359671853055602, -9.352894476754614, -9.348937477363904, -9.348359226622904, -9.345259603234013, -9.344056726904228, -9.343253852037947, -9.341990228180066, -9.34101372699871, -9.338546853445393, -9.336593727404477, -9.335445352056524, -9.331886728219558, -9.328775352237358, -9.327630853404344, -9.32624260262412, -9.324999976663946, -9.323584727358025, -9.32218560337526, -9.319421851741936, -9.318037102992719, -9.316833976996236, -9.313734352098493, -9.310685602020783, -9.308615227298162, -9.307414102003122, -9.303505851821399, -9.302292977138052, -9.301550728197697, -9.300863227008108, -9.299537103138903, -9.298849601601773, -9.298044976561243, -9.297581978073538, -9.297222299267844, -9.296343673717729, -9.296042642567613, -9.296091051176177, -9.296092078242282, -9.296425974962972, -9.296570406631076, -9.296571307988984, -9.297048307767454, -9.297239634523578, -9.297335656295331, -9.297241129332804, -9.297147018708301, -9.296815152401336, -9.29681598373287, -9.297197967608213, -9.297293981069856, -9.29719968253687, -9.296867213265045, -9.296534683626248, -9.295963748608074, -9.295488702688658, -9.295251588698363, -9.29539667501217, -9.295588735124227, -9.295638214086523, -9.295639602432573, -9.295783161478994, -9.296069816314798, -9.296356584526734, -9.295881282348205, -9.295739218618792, -9.295645004426774, -9.295122303138328, -9.294551977002243, -9.294029327032149, -9.293554126311506, -9.293031414276072, -9.292604451000898, -9.292605800897945, -9.292845033837011, -9.293274585186168, -9.293941854683169, -9.29399014689011, -9.293943583693363, -9.294563506539191, -9.295231161184972, -9.295660758479698, -9.296090574721404, -9.296424757789737, -9.296568780725352, -9.297236261001387, -9.297903685276367, -9.298570944263247, -9.299238202661286, -9.299857619609156, -9.300476927057161, -9.300763315724623, -9.301002135778276, -9.301479392257193, -9.301746977659832, -9.30195843341653, -9.30176902400359, -9.30124609484119, -9.301008789842967, -9.301105053539914, -9.300534175364305, -9.29996367018299, -9.299631806532204, -9.299871776194664, -9.300158938099798, -9.30039794641192, -9.3013046633086, -9.30230625092537, -9.30426100873433, -9.305262531785559, -9.306073250558029, -9.307265460498854, -9.307838522464806, -9.307982721614621, -9.307460509268992, -9.307652777783062, -9.308320149722306, -9.308892124204533, -9.309512578295426, -9.30994273922164, -9.310229776792994, -9.309754801916824, -9.309184279660712, -9.308376039712698, -9.307662990228948, -9.306616825241349, -9.305856339287743, -9.306000527297215, -9.306620691792943, -9.307145219779613, -9.30705070317049, -9.30690899803949, -9.307671971403977, -9.307815705688256, -9.30738816010961, -9.30677007355581, -9.306675879059355, -9.305629469089297, -9.305154192732914, -9.304821733978429, -9.304830602195409, -9.304544470841961, -9.303970692091752, -9.30291819876976, -9.301194790469594, -9.300237926182604, -9.299760526466741, -9.298900132705569, -9.2985181693669, -9.298136017522665, -9.296795322783566, -9.295742636828095, -9.29574412132763, -9.295553965721695, -9.294980541890084, -9.293927668202969, -9.293353874467869, -9.292971802593286, -9.291920115077119, -9.289815455799953, -9.28972045630286, -9.288955295916537, -9.288764749691438, -9.289244449452667, -9.289916309868147, -9.290780848613736, -9.291740732298706, -9.292509056729614, -9.292797443783513, -9.29366043007235, -9.295288689326071]))
type(maps.geometry)
geopandas.geoseries.GeoSeries
#Check unique values
maps['IDDIST'].is_unique
True
maps['IDDIST'].unique().size
1873
# Select only relevant columns
maps = maps[['IDDIST', 'geometry']]
maps = maps.rename({'IDDIST':'UBIGEO'}, axis =1 )
maps
| UBIGEO | geometry | |
|---|---|---|
| 0 | 100902 | POLYGON ((-75.31797 -9.29529, -75.31710 -9.297... |
| 1 | 100904 | POLYGON ((-74.64136 -8.82302, -74.64036 -8.828... |
| 2 | 250305 | POLYGON ((-75.02253 -8.74193, -75.02267 -8.742... |
| 3 | 250302 | POLYGON ((-75.13864 -8.56712, -75.13956 -8.569... |
| 4 | 250304 | POLYGON ((-75.01589 -8.44637, -75.01585 -8.446... |
| ... | ... | ... |
| 1868 | 100608 | POLYGON ((-76.08083 -9.13017, -76.08026 -9.130... |
| 1869 | 100609 | POLYGON ((-75.88828 -9.00906, -75.88756 -9.010... |
| 1870 | 100610 | POLYGON ((-75.91141 -8.88593, -75.91182 -8.886... |
| 1871 | 211105 | POLYGON ((-70.13203 -15.33382, -70.12355 -15.3... |
| 1872 | 150806 | POLYGON ((-77.37630 -10.89734, -77.37604 -10.8... |
1873 rows × 2 columns
maps.dtypes
UBIGEO object
geometry geometry
dtype: object
# Object or srting to int
maps['UBIGEO'] = maps['UBIGEO'].astype(str).astype(int)
maps
| UBIGEO | geometry | |
|---|---|---|
| 0 | 100902 | POLYGON ((-75.31797 -9.29529, -75.31710 -9.297... |
| 1 | 100904 | POLYGON ((-74.64136 -8.82302, -74.64036 -8.828... |
| 2 | 250305 | POLYGON ((-75.02253 -8.74193, -75.02267 -8.742... |
| 3 | 250302 | POLYGON ((-75.13864 -8.56712, -75.13956 -8.569... |
| 4 | 250304 | POLYGON ((-75.01589 -8.44637, -75.01585 -8.446... |
| ... | ... | ... |
| 1868 | 100608 | POLYGON ((-76.08083 -9.13017, -76.08026 -9.130... |
| 1869 | 100609 | POLYGON ((-75.88828 -9.00906, -75.88756 -9.010... |
| 1870 | 100610 | POLYGON ((-75.91141 -8.88593, -75.91182 -8.886... |
| 1871 | 211105 | POLYGON ((-70.13203 -15.33382, -70.12355 -15.3... |
| 1872 | 150806 | POLYGON ((-77.37630 -10.89734, -77.37604 -10.8... |
1873 rows × 2 columns
# WGS-84 (longitud-latitud projection)
maps.crs
#system of coordenates: CRS (due to where we are)
<Geographic 2D CRS: EPSG:4326>
Name: WGS 84
Axis Info [ellipsoidal]:
- Lat[north]: Geodetic latitude (degree)
- Lon[east]: Geodetic longitude (degree)
Area of Use:
- name: World
- bounds: (-180.0, -90.0, 180.0, 90.0)
Datum: World Geodetic System 1984
- Ellipsoid: WGS 84
- Prime Meridian: Greenwich
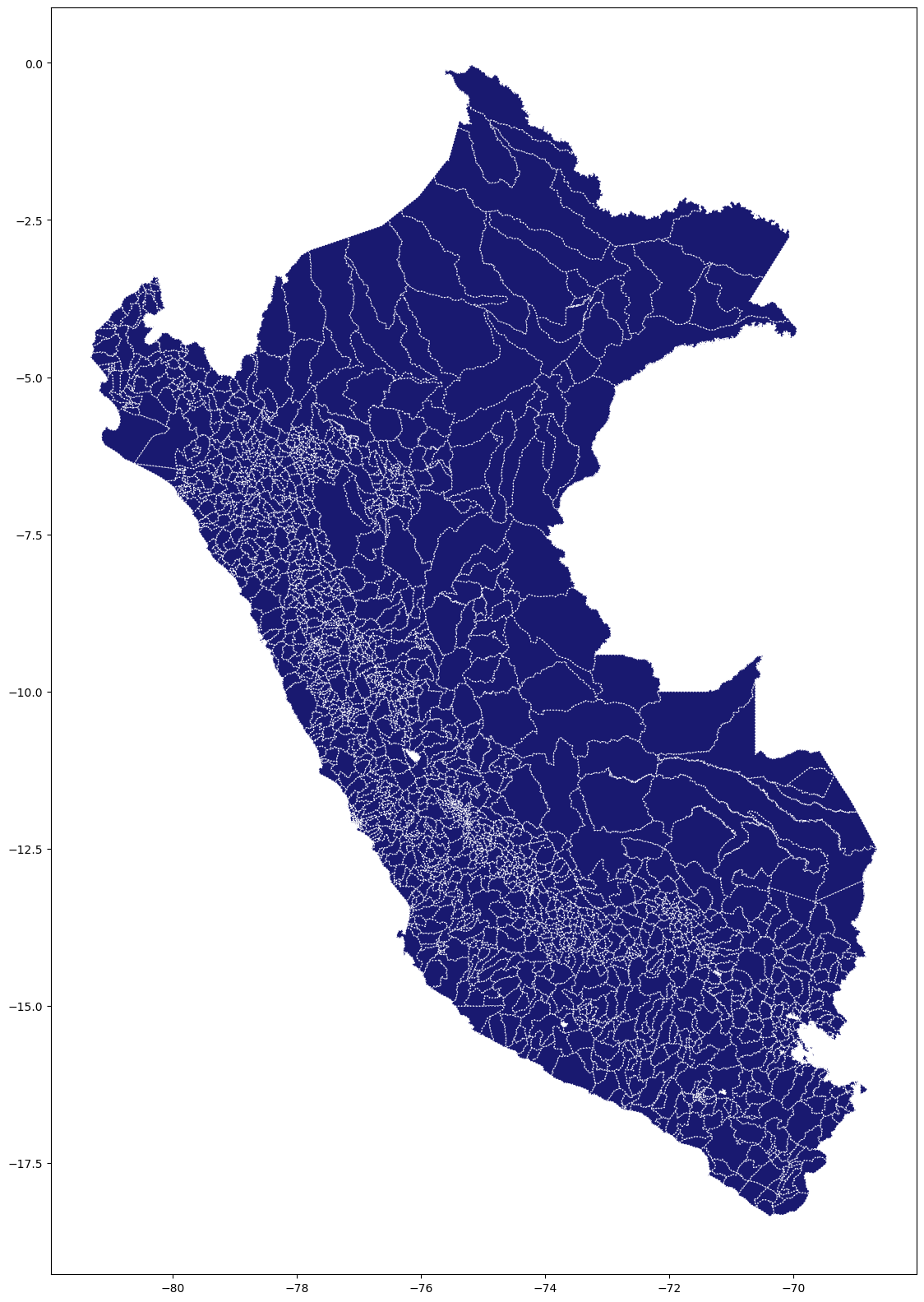
# Plot using geopandas
#figure and axis
fig, ax = plt.subplots(figsize=(20, 20))
maps.plot( ax=ax, color='midnightblue', linestyle='dotted',
edgecolor='white' )
plt.show()
Merge: Covid Data & Shapefile#
# Merge using inner way to drop missing values
dataset_cv = pd.merge(maps, cv_data, how="inner", on="UBIGEO")
dataset_cv
| UBIGEO | geometry | Departamento | Provincia | Distrito | Mes | Año | Casos | Muertes Covid | Muertes totales | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 100902 | POLYGON ((-75.31797 -9.29529, -75.31710 -9.297... | HUANUCO | PUERTO INCA | CODO DEL POZUZO | 3 | 2020 | 0 | 0 | 3 |
| 1 | 100902 | POLYGON ((-75.31797 -9.29529, -75.31710 -9.297... | HUANUCO | PUERTO INCA | CODO DEL POZUZO | 4 | 2020 | 0 | 1 | 2 |
| 2 | 100902 | POLYGON ((-75.31797 -9.29529, -75.31710 -9.297... | HUANUCO | PUERTO INCA | CODO DEL POZUZO | 5 | 2020 | 1 | 0 | 2 |
| 3 | 100902 | POLYGON ((-75.31797 -9.29529, -75.31710 -9.297... | HUANUCO | PUERTO INCA | CODO DEL POZUZO | 6 | 2020 | 7 | 0 | 1 |
| 4 | 100902 | POLYGON ((-75.31797 -9.29529, -75.31710 -9.297... | HUANUCO | PUERTO INCA | CODO DEL POZUZO | 7 | 2020 | 11 | 1 | 4 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 40360 | 150806 | POLYGON ((-77.37630 -10.89734, -77.37604 -10.8... | LIMA | HUAURA | HUAURA | 9 | 2021 | 9 | 0 | 20 |
| 40361 | 150806 | POLYGON ((-77.37630 -10.89734, -77.37604 -10.8... | LIMA | HUAURA | HUAURA | 10 | 2021 | 11 | 2 | 16 |
| 40362 | 150806 | POLYGON ((-77.37630 -10.89734, -77.37604 -10.8... | LIMA | HUAURA | HUAURA | 11 | 2021 | 28 | 1 | 10 |
| 40363 | 150806 | POLYGON ((-77.37630 -10.89734, -77.37604 -10.8... | LIMA | HUAURA | HUAURA | 12 | 2021 | 75 | 1 | 16 |
| 40364 | 150806 | POLYGON ((-77.37630 -10.89734, -77.37604 -10.8... | LIMA | HUAURA | HUAURA | 1 | 2022 | 39 | 0 | 7 |
40365 rows × 10 columns
# Function to filter dataset_covid by year, month and administrative space(province)
def subdata( a , b, c ):
if c == "All-Peru":
resultado = dataset_cv.loc[(dataset_cv['Año']== a) & (dataset_cv['Mes']== b)]
else:
resultado = dataset_cv.loc[(dataset_cv['Año']== a) & (dataset_cv['Mes']== b) & (dataset_cv['Provincia']== c)]
return resultado
base.Provincia.unique()
array(['PUERTO INCA', 'PADRE ABAD', 'CORONEL PORTILLO', 'UCAYALI',
'TRUJILLO', 'ASCOPE', 'OTUZCO', 'GRAN CHIMU', 'CONTUMAZA',
'PACASMAYO', 'TACNA', 'JORGE BASADRE', 'ILO', 'TARATA',
'CANDARAVE', 'MARISCAL NIETO', 'ISLAY', 'CHUCUITO',
'GENERAL SANCHEZ CERRO', 'CAMANA', 'AREQUIPA', 'EL COLLAO',
'YUNGUYO', 'CASTILLA', 'CAYLLOMA', 'PUNO', 'CONDESUYOS',
'CARAVELI', 'SAN ROMAN', 'LAMPA', 'MOHO', 'HUANCANE', 'CHINCHEROS',
'HUAMALIES', 'SANTA', 'ANTONIO RAYMONDI', 'YUNGAY', 'HUAYLAS',
'CARLOS FERMIN FITZCARRALD', 'CORONGO', 'POMABAMBA', 'MELGAR',
'ESPINAR', 'AZANGARO', 'NASCA', 'SAN ANTONIO DE PUTINA',
'LA UNI<d2>N', 'YAROWILCA', 'HUARAZ', 'RECUAY', 'DOS DE MAYO',
'LAURICOCHA', 'BOLOGNESI', 'AMBO', 'CAJAMARCA', 'BOLIVAR',
'CHEPEN', 'MARISCAL CACERES', 'SAN MARCOS', 'HUALLAGA', 'CHICLAYO',
'CELENDIN', 'OCROS', 'HUARI', 'MARA<d1>ON', 'HUAYTARA', 'CARABAYA',
'QUISPICANCHI', 'PISCO', 'ANDAHUAYLAS', 'ASUNCION', 'SIHUAS',
'PALLASCA', 'HUARMEY', 'AIJA', 'HUANUCO', 'PACHITEA', 'OXAPAMPA',
'PICOTA', 'CHACHAPOYAS', 'CHOTA', 'CUTERVO', 'EL DORADO', 'LUYA',
'RODRIGUEZ DE MENDOZA', 'PASCO', 'DANIEL ALCIDES CARRION',
'CAJATAMBO', 'JAEN', 'LAMAS', 'LAMBAYEQUE', 'FERRE<d1>AFE',
'SAN MARTIN', 'CHURCAMPA', 'TAYACAJA', 'CASMA', 'CAJABAMBA',
'YAUYOS', 'HUANCAVELICA', 'CALCA', 'CASTROVIRREYNA', 'HUAROCHIRI',
'CHUMBIVILCAS', 'CANAS', 'AYMARAES', 'GRAU', 'CANCHIS',
'COTABAMBAS', 'ACOBAMBA', 'CA<d1>ETE', 'LIMA', 'CHINCHA',
'URUBAMBA', 'CARHUAZ', 'HUANCAYO', 'LA CONVENCION', 'JAUJA',
'PARURO', 'CONCEPCION', 'SANDIA', 'ICA', 'ANTABAMBA', 'TARMA',
'HUARAL', 'TAHUAMANU', 'CHANCHAMAYO', 'HUAURA', 'YAULI',
'TAMBOPATA', 'HUANTA', 'LA MAR', 'SANTA CRUZ', 'HUALGAYOC',
'SANCHEZ CARRION', 'UTCUBAMBA', 'SAN IGNACIO', 'CUSCO', 'CHUPACA',
'PROV. CONST. DEL CALLAO', 'MOYOBAMBA', 'MAYNAS', 'TUMBES',
'CONTRALMIRANTE VILLAR', 'BAGUA', 'ZARUMILLA', 'BONGARA', 'RIOJA',
'REQUENA', 'CONDORCANQUI', 'DATEM DEL MARA<d1>ON',
'MARISCAL RAMON CASTILLA', 'LORETO', 'SECHURA', 'HUANCABAMBA',
'PIURA', 'MORROPON', 'BELLAVISTA', 'HUACAYBAMBA',
'MARISCAL LUZURIAGA', 'PUTUMAYO', 'SATIPO', 'CANTA', 'OYON',
'ATALAYA', 'JUNIN', 'PATAZ', 'BARRANCA', 'ALTO AMAZONAS', 'JULCAN',
'SANTIAGO DE CHUCO', 'VIRU', 'TOCACHE', 'SAN MIGUEL', 'PALPA',
'ACOMAYO', 'ABANCAY', 'ANGARAES', 'MANU', 'PAUCARTAMBO', 'ANTA',
'LUCANAS', 'SUCRE', 'HUANCA SANCOS', 'VILCAS HUAMAN',
'VICTOR FAJARDO', 'SAN PABLO', 'PURUS', 'SULLANA', 'PAITA',
'AYABACA', 'TALARA', 'PARINACOCHAS', 'P<c0>UCAR DEL SARA SARA',
'CANGALLO', 'HUAMANGA', 'LEONCIO PRADO'], dtype=object)
#Second wave
base = subdata( 2020 , 8, "All-Peru") # all peru Aug-2020
#Second wave only Lima
base_lima = subdata( 2020 , 8, "LIMA") # Lima Aug-2020
#Second wave only Lima
base_ayacucho = subdata( 2020 , 10, "LUCANAS") # Lima Aug-2020
base_ayacucho
| UBIGEO | geometry | Departamento | Provincia | Distrito | Mes | Año | Casos | Muertes Covid | Muertes totales | |
|---|---|---|---|---|---|---|---|---|---|---|
| 24617 | 50618 | POLYGON ((-74.45485 -14.15098, -74.45421 -14.1... | AYACUCHO | LUCANAS | SAN PEDRO DE PALCO | 10 | 2020 | 0 | 0 | 0 |
| 37773 | 50617 | POLYGON ((-74.12341 -14.73414, -74.12249 -14.7... | AYACUCHO | LUCANAS | SAN PEDRO | 10 | 2020 | 1 | 0 | 0 |
| 37819 | 50621 | POLYGON ((-74.39258 -14.72377, -74.39211 -14.7... | AYACUCHO | LUCANAS | SANTA LUCIA | 10 | 2020 | 0 | 0 | 0 |
| 37842 | 50615 | POLYGON ((-74.28449 -14.69725, -74.28211 -14.6... | AYACUCHO | LUCANAS | SAN CRISTOBAL | 10 | 2020 | 2 | 0 | 0 |
| 37957 | 50619 | POLYGON ((-74.04573 -14.91000, -74.04615 -14.9... | AYACUCHO | LUCANAS | SANCOS | 10 | 2020 | 6 | 0 | 0 |
| 38003 | 50614 | POLYGON ((-74.36156 -14.77078, -74.35963 -14.7... | AYACUCHO | LUCANAS | SAISA | 10 | 2020 | 0 | 0 | 0 |
| 38026 | 50605 | POLYGON ((-73.87660 -14.80292, -73.87742 -14.8... | AYACUCHO | LUCANAS | CHAVI<d1>A | 10 | 2020 | 0 | 0 | 2 |
| 38049 | 50616 | POLYGON ((-74.17274 -14.59704, -74.17246 -14.5... | AYACUCHO | LUCANAS | SAN JUAN | 10 | 2020 | 0 | 0 | 0 |
| 38417 | 50609 | POLYGON ((-74.43725 -14.52504, -74.43454 -14.5... | AYACUCHO | LUCANAS | LEONCIO PRADO | 10 | 2020 | 1 | 0 | 0 |
| 38440 | 50606 | POLYGON ((-73.85391 -14.27810, -73.85018 -14.2... | AYACUCHO | LUCANAS | CHIPAO | 10 | 2020 | 2 | 0 | 1 |
| 38463 | 50611 | POLYGON ((-74.27602 -14.27152, -74.27477 -14.2... | AYACUCHO | LUCANAS | LUCANAS | 10 | 2020 | 0 | 0 | 2 |
| 38486 | 50603 | POLYGON ((-73.89004 -14.26761, -73.88869 -14.2... | AYACUCHO | LUCANAS | CABANA | 10 | 2020 | 0 | 0 | 0 |
| 38509 | 50612 | POLYGON ((-74.64219 -14.23868, -74.64053 -14.2... | AYACUCHO | LUCANAS | OCA<d1>A | 10 | 2020 | 9 | 1 | 2 |
| 38532 | 50613 | POLYGON ((-74.49890 -14.38808, -74.49748 -14.3... | AYACUCHO | LUCANAS | OTOCA | 10 | 2020 | 0 | 0 | 0 |
| 38555 | 50604 | POLYGON ((-73.93008 -14.32350, -73.92803 -14.3... | AYACUCHO | LUCANAS | CARMEN SALCEDO | 10 | 2020 | 2 | 0 | 2 |
| 38624 | 50608 | POLYGON ((-74.67672 -14.07173, -74.67674 -14.0... | AYACUCHO | LUCANAS | LARAMATE | 10 | 2020 | 0 | 0 | 0 |
| 38647 | 50610 | POLYGON ((-74.85561 -14.07523, -74.85274 -14.0... | AYACUCHO | LUCANAS | LLAUTA | 10 | 2020 | 0 | 0 | 0 |
| 38716 | 50602 | POLYGON ((-74.06654 -14.02992, -74.06581 -14.0... | AYACUCHO | LUCANAS | AUCARA | 10 | 2020 | 5 | 1 | 2 |
| 38785 | 50601 | POLYGON ((-74.09914 -14.51889, -74.09729 -14.5... | AYACUCHO | LUCANAS | PUQUIO | 10 | 2020 | 46 | 1 | 1 |
| 38808 | 50620 | POLYGON ((-73.94270 -14.19863, -73.94250 -14.1... | AYACUCHO | LUCANAS | SANTA ANA DE HUAYCAHUACHO | 10 | 2020 | 0 | 0 | 0 |
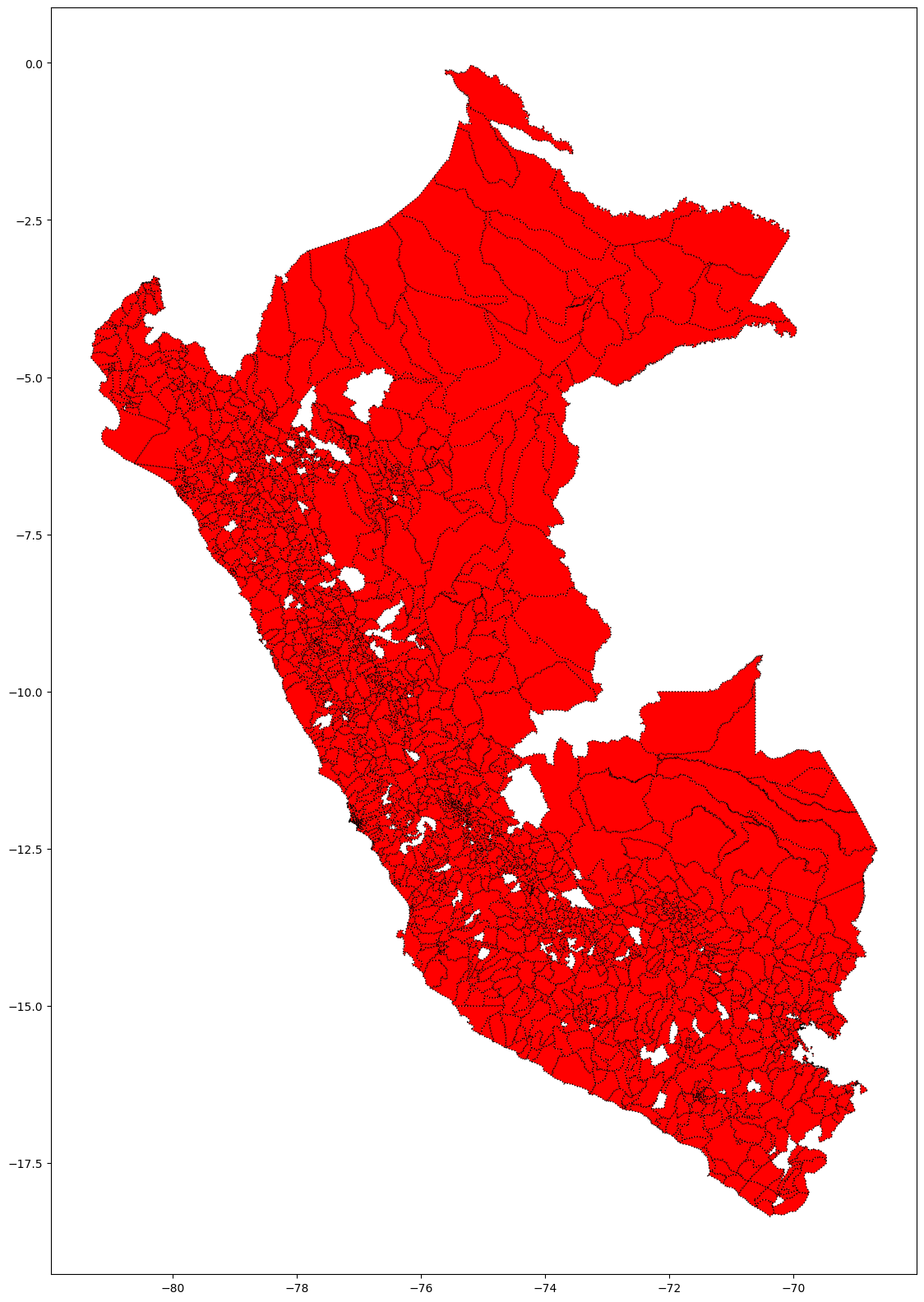
fig, ax = plt.subplots(figsize=(20, 20))
base.plot( ax=ax, color='red', linestyle='dotted',
edgecolor='black' )
plt.show()
#does not picture the provinces who dont have covid cases.
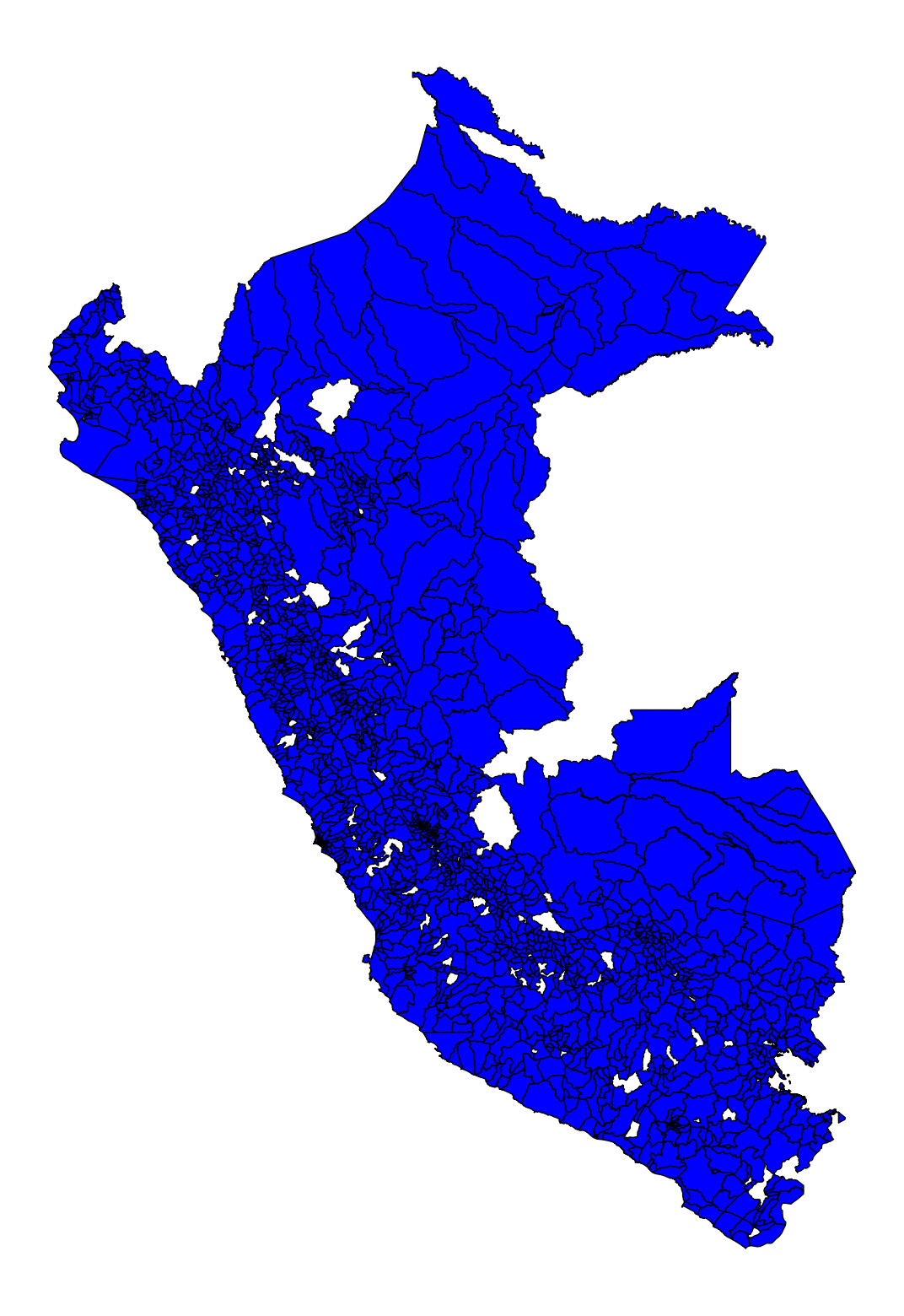
# Plot without axis
fig, ax = plt.subplots(figsize=(20, 20))
base.plot(ax=ax,color='blue', linestyle='-',
edgecolor='black')
ax.set_axis_off()
Plot covid infection at disctrict level#
# First always check the distribution
fig, ax = plt.subplots(figsize=(10, 10))
base["Casos"].hist(bins = 100)
#check the distribution of the variables BEFORE PLOTTING
<AxesSubplot:>
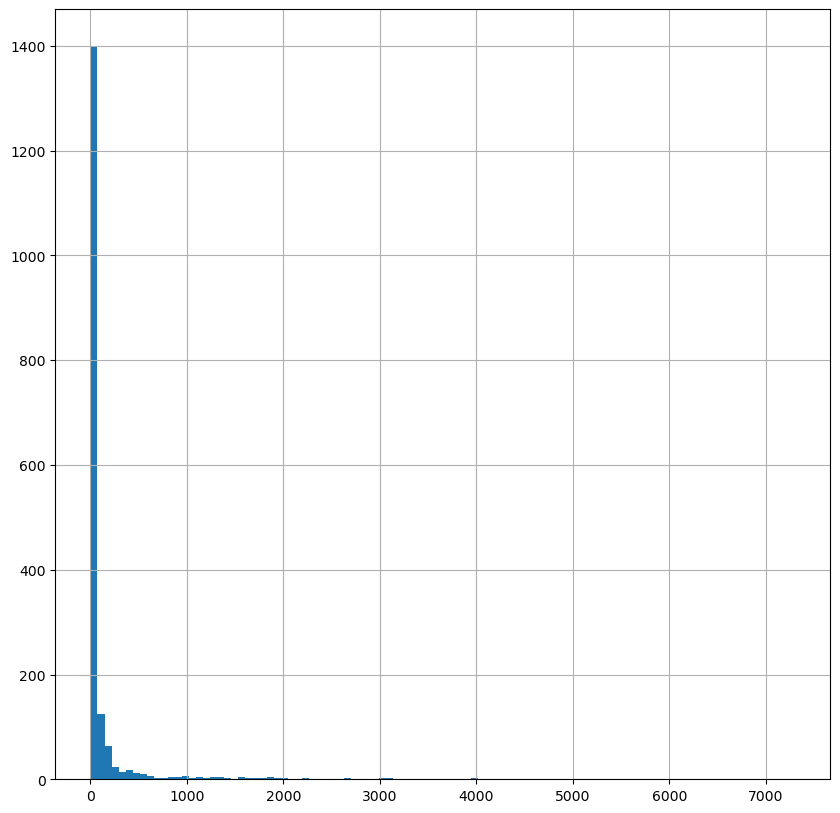
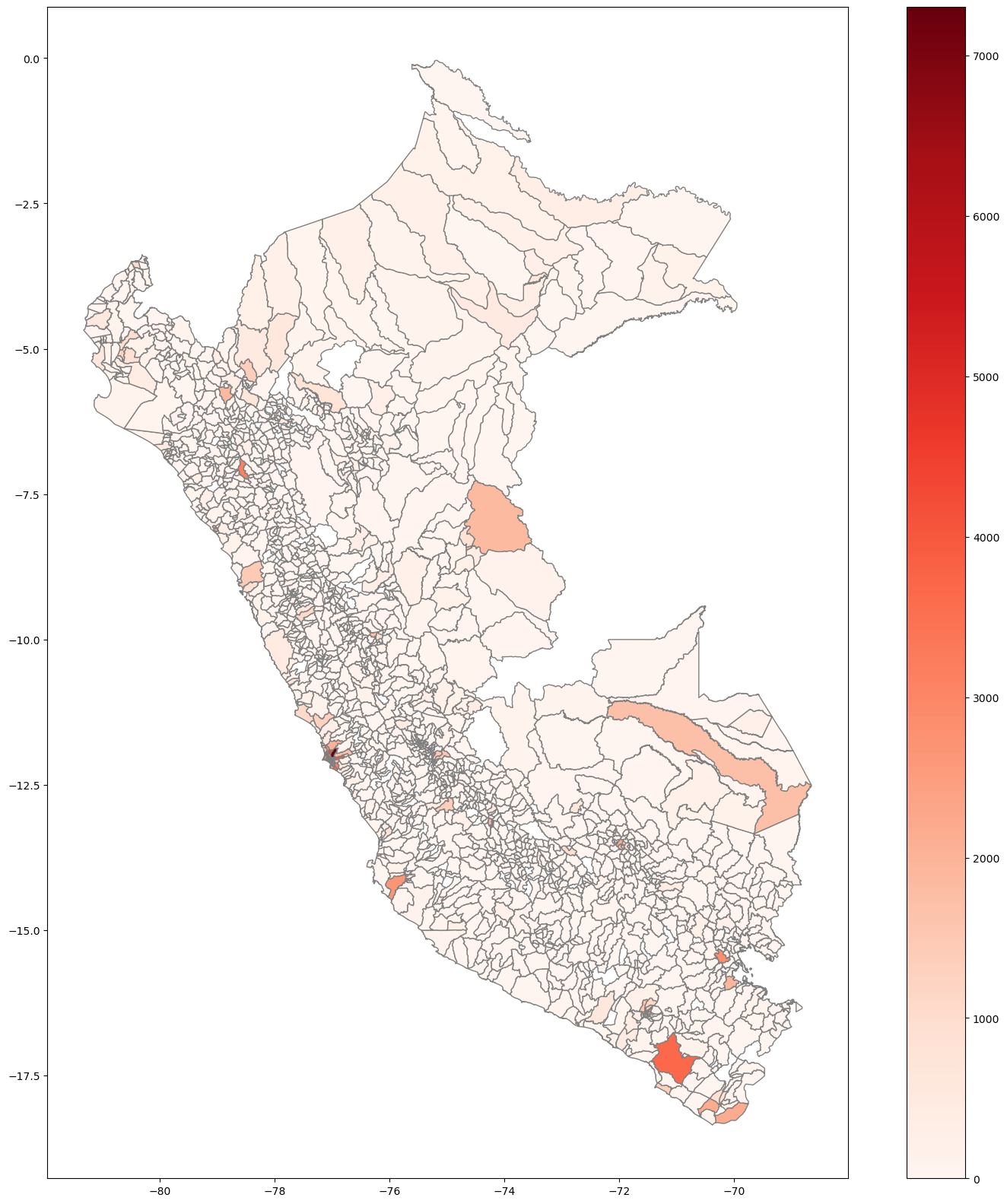
# Plot covid infection at disctrict level
base.plot( column='Casos', cmap='Reds',
figsize=(20, 20),
linestyle='-',
edgecolor='gray',
legend = True )
<AxesSubplot:>
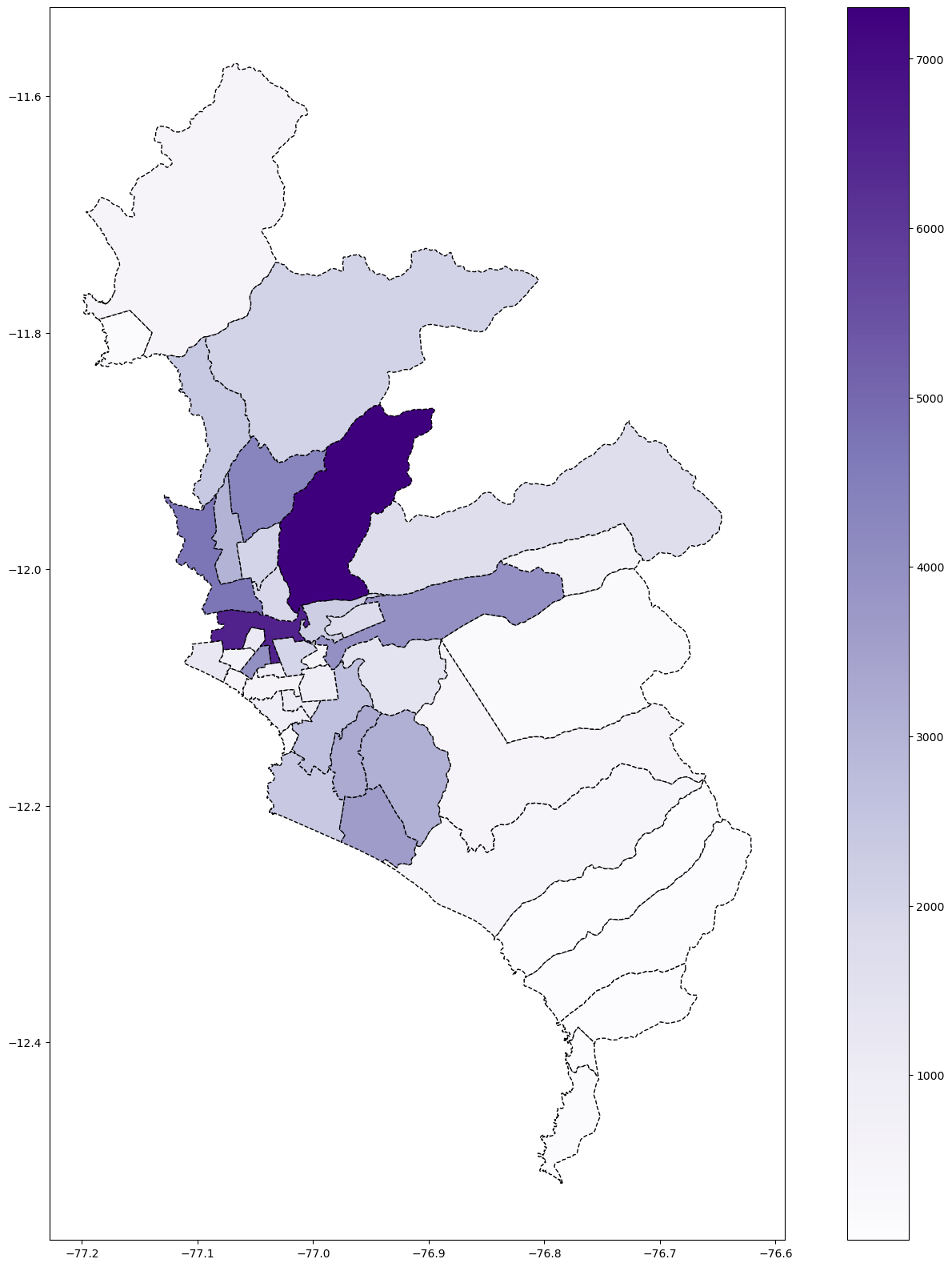
# Covid & Lima Matropolitana
base_lima.plot(column='Casos', cmap='Purples', figsize=(20, 20), linestyle='--',
edgecolor='black',
legend = True)
# save map
plt.savefig(r'Lima_covid_Heatmap.png')
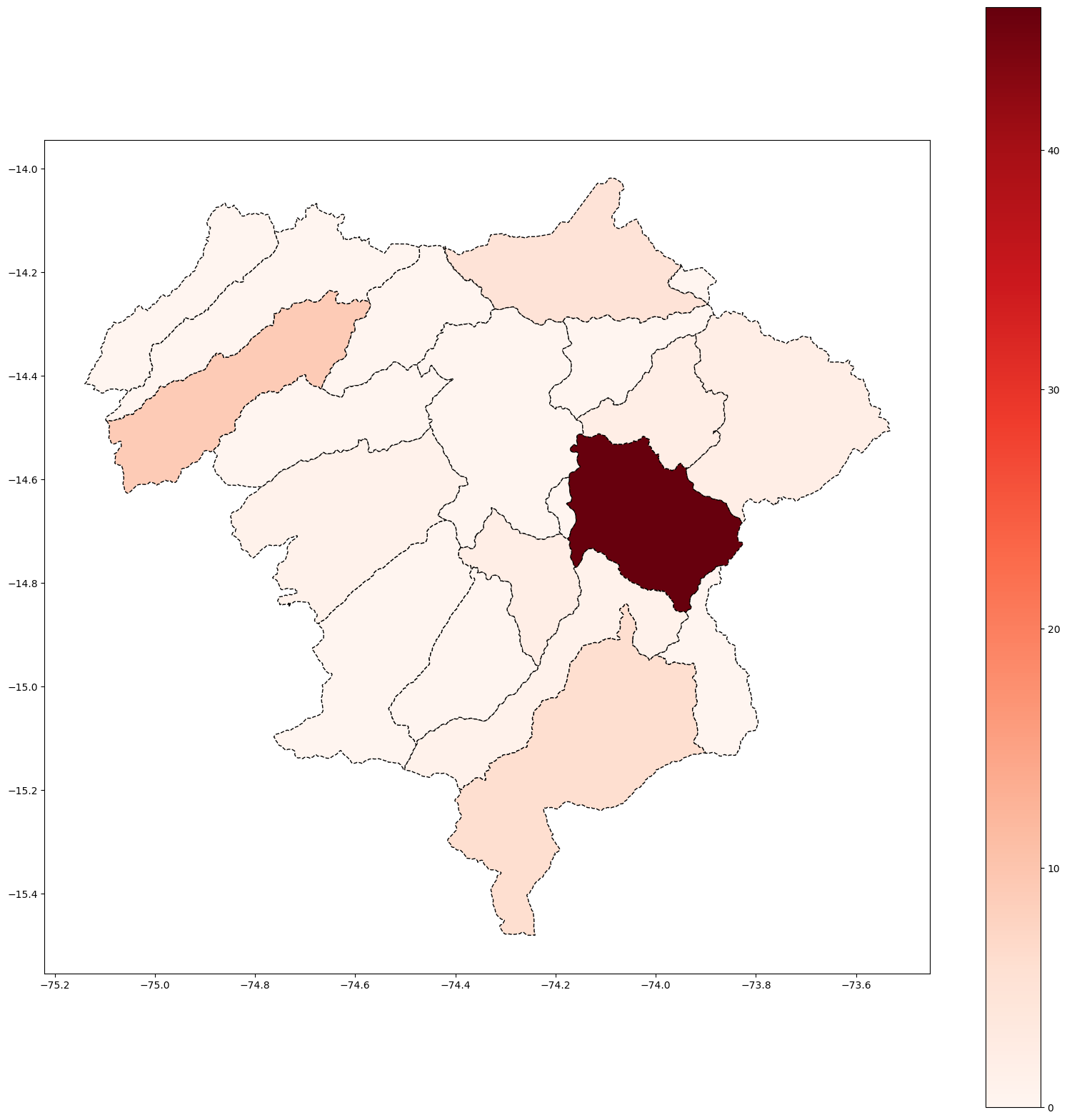
# Covid & Lima Matropolitana
base_ayacucho.plot(column='Casos', cmap='Reds', figsize=(20, 20), linestyle='--',
edgecolor='black',
legend = True)
# save map
plt.savefig(r'Ayacucho_covid_Heatmap.png')
base_lima.crs
<Geographic 2D CRS: EPSG:4326>
Name: WGS 84
Axis Info [ellipsoidal]:
- Lat[north]: Geodetic latitude (degree)
- Lon[east]: Geodetic longitude (degree)
Area of Use:
- name: World
- bounds: (-180.0, -90.0, 180.0, 90.0)
Datum: World Geodetic System 1984
- Ellipsoid: WGS 84
- Prime Meridian: Greenwich
Get Centroid#
# Centroid (point in the center)
#base_lima = base_lima.to_crs("EPSG:32733")
#WGS84 (MAP IN CURVE) > PROJ IN METER "EPSG:24891" TO CALCULATE CENTROID > CONVERT TO CURVE AGAIN (THE CENTROID CAN BE OUT BECAUSE IF THE IMAGE IS CONCAVA)
base_lima['centroid'] = base_lima['geometry'].to_crs("EPSG:24891").centroid.to_crs("EPSG:4326")
C:\Users\DELL\anaconda3\lib\site-packages\geopandas\geodataframe.py:1443: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
super().__setitem__(key, value)
base_lima
| UBIGEO | geometry | Departamento | Provincia | Distrito | Mes | Año | Casos | Muertes Covid | Muertes totales | centroid | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 8147 | 150119 | POLYGON ((-76.70549 -12.17672, -76.70429 -12.1... | LIMA | LIMA | LURIN | 8 | 2020 | 452 | 29 | 68 | POINT (-76.80109 -12.23426) |
| 9090 | 150102 | POLYGON ((-77.06517 -11.57512, -77.06505 -11.5... | LIMA | LIMA | ANCON | 8 | 2020 | 460 | 30 | 51 | POINT (-77.09580 -11.70252) |
| 11689 | 150131 | POLYGON ((-77.04859 -12.08504, -77.04762 -12.0... | LIMA | LIMA | SAN ISIDRO | 8 | 2020 | 551 | 30 | 86 | POINT (-77.03667 -12.09898) |
| 11712 | 150120 | POLYGON ((-77.07210 -12.08358, -77.06348 -12.0... | LIMA | LIMA | MAGDALENA DEL MAR | 8 | 2020 | 478 | 49 | 94 | POINT (-77.06669 -12.09384) |
| 11735 | 150130 | POLYGON ((-76.98732 -12.07994, -76.98425 -12.0... | LIMA | LIMA | SAN BORJA | 8 | 2020 | 1012 | 54 | 123 | POINT (-76.99521 -12.09755) |
| 11758 | 150116 | POLYGON ((-77.02807 -12.07871, -77.02435 -12.0... | LIMA | LIMA | LINCE | 8 | 2020 | 855 | 76 | 129 | POINT (-77.03567 -12.08579) |
| 11781 | 150140 | POLYGON ((-76.95923 -12.07576, -76.95831 -12.0... | LIMA | LIMA | SANTIAGO DE SURCO | 8 | 2020 | 2628 | 171 | 394 | POINT (-76.98431 -12.12699) |
| 11850 | 150121 | POLYGON ((-77.04966 -12.07025, -77.05097 -12.0... | LIMA | LIMA | PUEBLO LIBRE | 8 | 2020 | 633 | 61 | 140 | POINT (-77.06574 -12.07508) |
| 12379 | 150139 | POLYGON ((-77.14355 -11.79587, -77.14061 -11.7... | LIMA | LIMA | SANTA ROSA | 8 | 2020 | 114 | 9 | 15 | POINT (-77.16325 -11.80501) |
| 15047 | 150106 | POLYGON ((-76.89995 -11.72979, -76.89921 -11.7... | LIMA | LIMA | CARABAYLLO | 8 | 2020 | 2054 | 183 | 281 | POINT (-76.97291 -11.80738) |
| 21579 | 150113 | POLYGON ((-77.03811 -12.06768, -77.03629 -12.0... | LIMA | LIMA | JESUS MARIA | 8 | 2020 | 4012 | 147 | 186 | POINT (-77.04821 -12.07811) |
| 22131 | 150124 | POLYGON ((-76.77367 -12.42553, -76.77332 -12.4... | LIMA | LIMA | PUCUSANA | 8 | 2020 | 90 | 8 | 13 | POINT (-76.77562 -12.46584) |
| 22154 | 150138 | MULTIPOLYGON (((-76.78117 -12.40917, -76.78144... | LIMA | LIMA | SANTA MARIA DEL MAR | 8 | 2020 | 26 | 0 | 0 | POINT (-76.76740 -12.40858) |
| 22315 | 150108 | POLYGON ((-77.00745 -12.16033, -77.00781 -12.1... | LIMA | LIMA | CHORRILLOS | 8 | 2020 | 2400 | 203 | 333 | POINT (-77.00589 -12.19265) |
| 22384 | 150104 | POLYGON ((-77.01945 -12.13060, -77.01915 -12.1... | LIMA | LIMA | BARRANCO | 8 | 2020 | 298 | 26 | 62 | POINT (-77.02086 -12.14403) |
| 22430 | 150143 | POLYGON ((-76.91078 -12.12233, -76.91191 -12.1... | LIMA | LIMA | VILLA MARIA DEL TRIUNFO | 8 | 2020 | 3123 | 324 | 508 | POINT (-76.91899 -12.17039) |
| 22453 | 150133 | POLYGON ((-76.95434 -12.11556, -76.95416 -12.1... | LIMA | LIMA | SAN JUAN DE MIRAFLORES | 8 | 2020 | 3251 | 267 | 479 | POINT (-76.96651 -12.15786) |
| 22568 | 150122 | POLYGON ((-77.02656 -12.10820, -77.02631 -12.1... | LIMA | LIMA | MIRAFLORES | 8 | 2020 | 951 | 66 | 178 | POINT (-77.02893 -12.12091) |
| 22614 | 150141 | POLYGON ((-77.01498 -12.10778, -77.01469 -12.1... | LIMA | LIMA | SURQUILLO | 8 | 2020 | 1103 | 91 | 170 | POINT (-77.01299 -12.11344) |
| 23925 | 150127 | POLYGON ((-76.64078 -12.21377, -76.64000 -12.2... | LIMA | LIMA | PUNTA NEGRA | 8 | 2020 | 54 | 3 | 7 | POINT (-76.70943 -12.30363) |
| 24086 | 150134 | POLYGON ((-76.99780 -12.06084, -76.99810 -12.0... | LIMA | LIMA | SAN LUIS | 8 | 2020 | 567 | 39 | 72 | POINT (-76.99724 -12.07409) |
| 24109 | 150136 | POLYGON ((-77.07857 -12.06228, -77.07815 -12.0... | LIMA | LIMA | SAN MIGUEL | 8 | 2020 | 1205 | 89 | 190 | POINT (-77.09007 -12.07644) |
| 24546 | 150142 | POLYGON ((-76.94232 -12.18247, -76.93731 -12.1... | LIMA | LIMA | VILLA EL SALVADOR | 8 | 2020 | 3625 | 346 | 415 | POINT (-76.94528 -12.21928) |
| 24569 | 150126 | POLYGON ((-76.65267 -12.19256, -76.65207 -12.1... | LIMA | LIMA | PUNTA HERMOSA | 8 | 2020 | 35 | 1 | 7 | POINT (-76.74377 -12.26927) |
| 25857 | 150129 | POLYGON ((-76.67773 -12.33800, -76.67787 -12.3... | LIMA | LIMA | SAN BARTOLO | 8 | 2020 | 80 | 4 | 10 | POINT (-76.72414 -12.36938) |
| 26271 | 150123 | POLYGON ((-76.88134 -12.07220, -76.86137 -12.1... | LIMA | LIMA | PACHACAMAC | 8 | 2020 | 464 | 34 | 76 | POINT (-76.80961 -12.15999) |
| 26317 | 150115 | POLYGON ((-77.01661 -12.05889, -77.01559 -12.0... | LIMA | LIMA | LA VICTORIA | 8 | 2020 | 2003 | 211 | 351 | POINT (-77.01739 -12.07233) |
| 26340 | 150114 | POLYGON ((-76.94006 -12.06405, -76.93942 -12.0... | LIMA | LIMA | LA MOLINA | 8 | 2020 | 1406 | 81 | 154 | POINT (-76.92573 -12.08809) |
| 26386 | 150105 | POLYGON ((-77.04206 -12.05348, -77.04198 -12.0... | LIMA | LIMA | BRE<d1>A | 8 | 2020 | 1129 | 95 | 180 | POINT (-77.05059 -12.05916) |
| 26593 | 150137 | POLYGON ((-76.93981 -12.03913, -76.93796 -12.0... | LIMA | LIMA | SANTA ANITA | 8 | 2020 | 1762 | 169 | 261 | POINT (-76.96314 -12.04321) |
| 26662 | 150109 | POLYGON ((-76.72948 -11.99943, -76.72596 -11.9... | LIMA | LIMA | CIENEGUILLA | 8 | 2020 | 161 | 12 | 25 | POINT (-76.77752 -12.07646) |
| 27743 | 150101 | POLYGON ((-77.00474 -12.04166, -77.00296 -12.0... | LIMA | LIMA | LIMA | 8 | 2020 | 6510 | 658 | 666 | POINT (-77.04887 -12.05103) |
| 27766 | 150111 | POLYGON ((-76.94408 -12.02075, -76.94348 -12.0... | LIMA | LIMA | EL AGUSTINO | 8 | 2020 | 2248 | 153 | 209 | POINT (-76.98740 -12.04010) |
| 27789 | 150128 | POLYGON ((-77.02034 -12.01783, -77.02032 -12.0... | LIMA | LIMA | RIMAC | 8 | 2020 | 1911 | 163 | 277 | POINT (-77.03268 -12.02358) |
| 27812 | 150103 | POLYGON ((-76.83508 -11.99627, -76.83467 -11.9... | LIMA | LIMA | ATE | 8 | 2020 | 3969 | 382 | 545 | POINT (-76.87747 -12.03257) |
| 27835 | 150107 | POLYGON ((-76.72918 -11.96426, -76.72809 -11.9... | LIMA | LIMA | CHACLACAYO | 8 | 2020 | 512 | 31 | 62 | POINT (-76.76695 -11.99164) |
| 27858 | 150112 | POLYGON ((-77.02858 -11.96462, -77.02794 -11.9... | LIMA | LIMA | INDEPENDENCIA | 8 | 2020 | 2044 | 181 | 277 | POINT (-77.04604 -11.98747) |
| 27904 | 150135 | POLYGON ((-77.08576 -11.94869, -77.08557 -11.9... | LIMA | LIMA | SAN MARTIN DE PORRES | 8 | 2020 | 4730 | 498 | 768 | POINT (-77.08841 -11.99283) |
| 27927 | 150117 | POLYGON ((-77.07044 -11.95013, -77.07008 -11.9... | LIMA | LIMA | LOS OLIVOS | 8 | 2020 | 3023 | 279 | 395 | POINT (-77.07437 -11.97242) |
| 27950 | 150110 | POLYGON ((-77.04639 -11.89743, -77.04551 -11.8... | LIMA | LIMA | COMAS | 8 | 2020 | 4323 | 533 | 753 | POINT (-77.03890 -11.92906) |
| 27973 | 150118 | POLYGON ((-76.71091 -11.89215, -76.70963 -11.8... | LIMA | LIMA | LURIGANCHO | 8 | 2020 | 1669 | 103 | 172 | POINT (-76.80130 -11.95849) |
| 27996 | 150132 | POLYGON ((-76.92923 -11.87063, -76.92891 -11.8... | LIMA | LIMA | SAN JUAN DE LURIGANCHO | 8 | 2020 | 7303 | 729 | 1071 | POINT (-76.97149 -11.94596) |
| 28042 | 150125 | POLYGON ((-77.09076 -11.81966, -77.09066 -11.8... | LIMA | LIMA | PUENTE PIEDRA | 8 | 2020 | 2437 | 189 | 311 | POINT (-77.08665 -11.87212) |
# fig, ax = plt.subplots(figsize=(20, 20))
# # Heatmap Lima and Covid infection
# base_lima.plot(column='Casos', cmap='Reds', linestyle='--',
# edgecolor='black',
# legend = True, ax=ax)
props = dict(boxstyle='round', facecolor = 'linen', alpha =1)
props
{'boxstyle': 'round', 'facecolor': 'linen', 'alpha': 1}
# for point in base_lima.iterrows():
# print(point[1]['Distrito'])
Add District Names in Centroid#
fig, ax = plt.subplots(figsize=(20, 20))
# Heatmap Lima and Covid infection
base_lima.plot(column='Casos', cmap='Reds', linestyle='--',
edgecolor='black',
legend = True, ax=ax)
<AxesSubplot:>
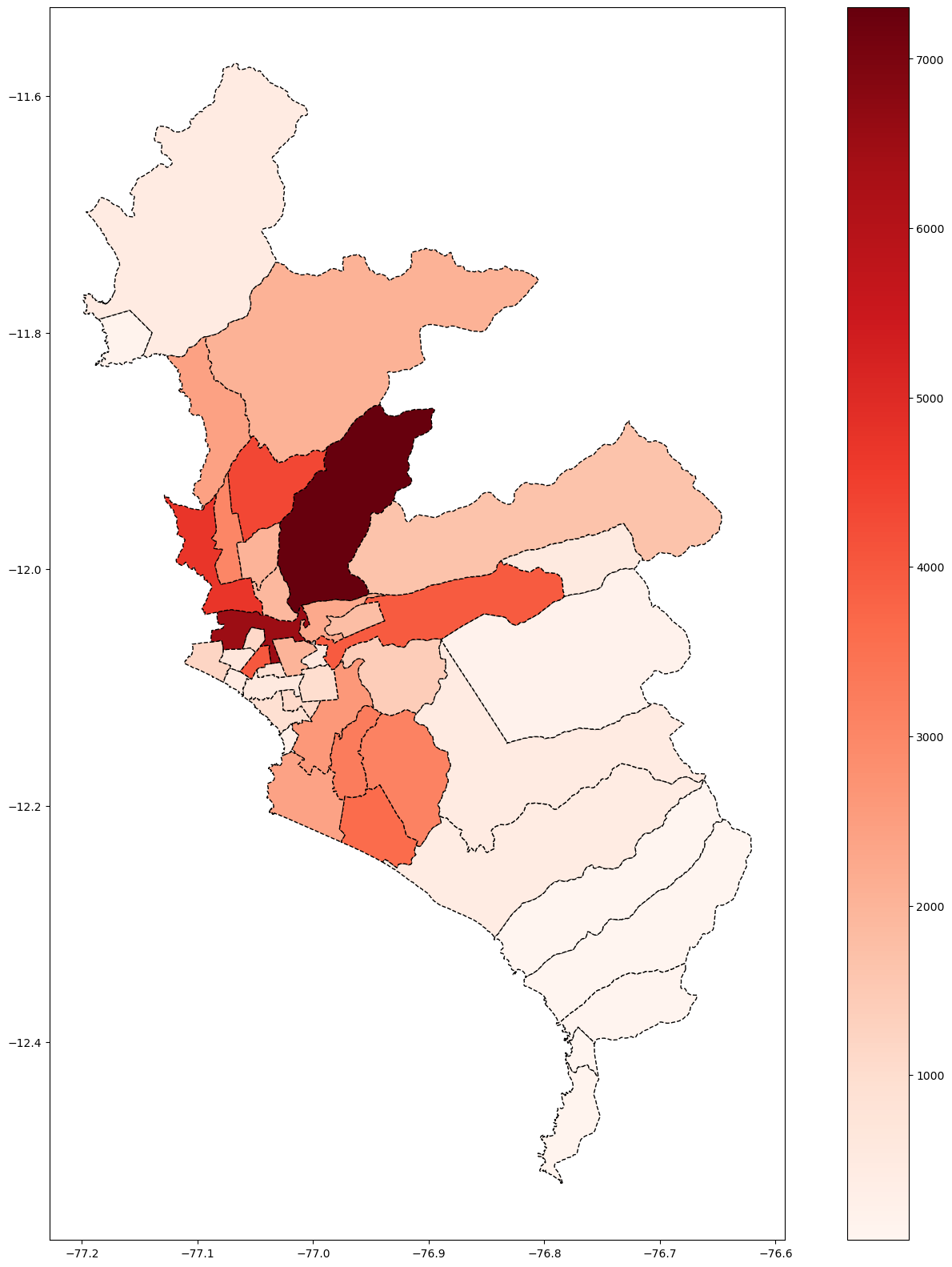
#base_lima['centroid']
# for point in base_lima.iterrows():
# print(point[1]['Distrito'])
props = dict(boxstyle='round', facecolor = 'linen', alpha =1)
props
{'boxstyle': 'round', 'facecolor': 'linen', 'alpha': 1}
fig, ax = plt.subplots(figsize=(20, 20))
# Heatmap Lima and Covid infection
base_lima.plot(column='Casos', cmap='Reds', linestyle='--',
edgecolor='black',
legend = True, ax=ax)
# add district name in litle box
props = dict(boxstyle='round', facecolor = 'linen', alpha =1)
for point in base_lima.iterrows():
ax.text( point[1]['centroid'].x,
point[1]['centroid'].y,
point[1]['Distrito'],
horizontalalignment='center',
fontsize=10,
bbox=props )
plt.savefig(r'Lima_covid_Heatmap_2.png')
#plot.savefig(r'Lima_covid_Heatmap_2.png')
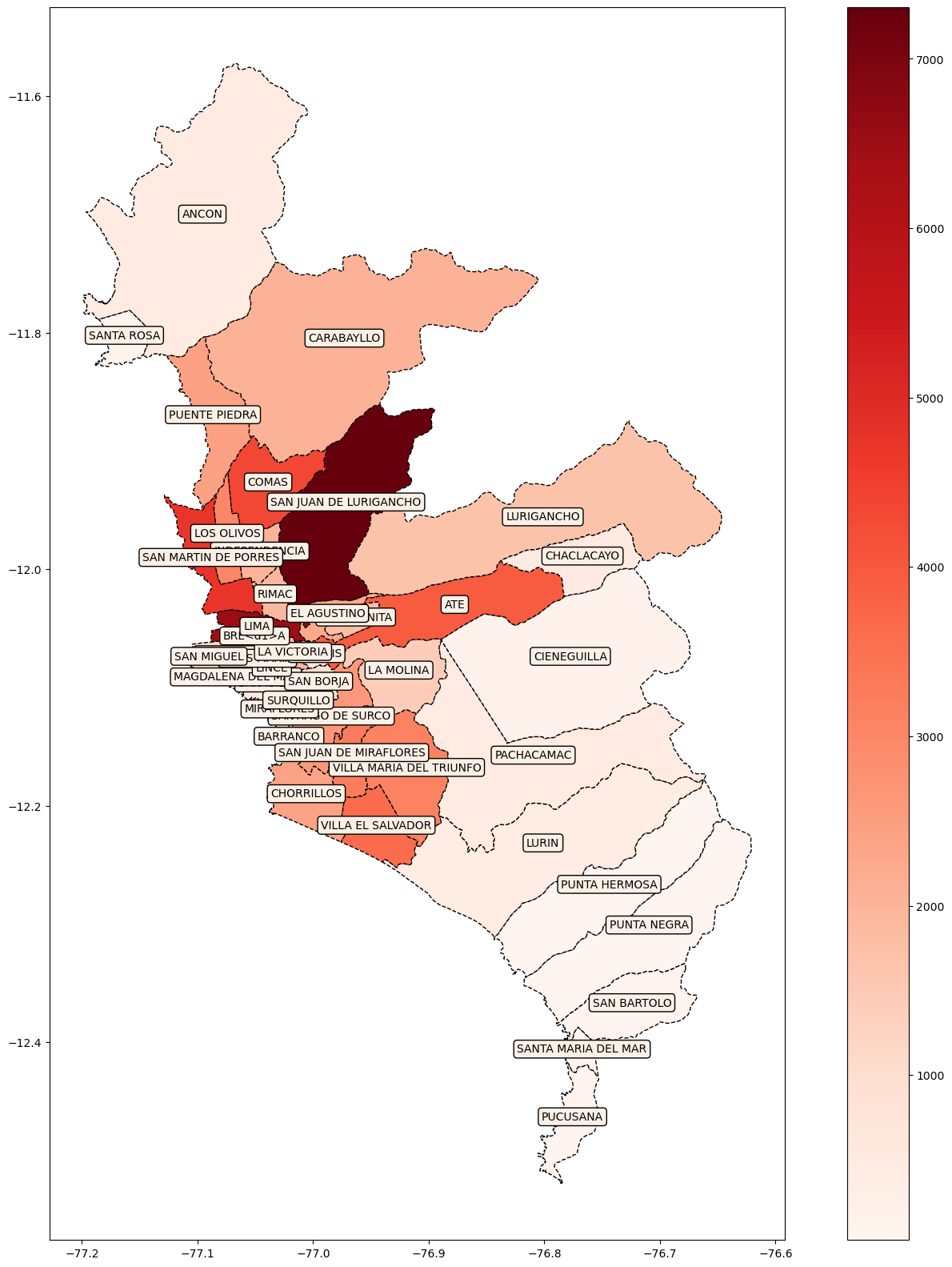
# Using legend by district
leg_kwds={'title':'District Name',
'loc': 'upper left',
'bbox_to_anchor':(1, 1.03),
'ncol':2}
base_lima.plot(column='Distrito', cmap='tab20c', figsize=(20, 20), legend = True, linestyle='-',
edgecolor='black',
legend_kwds = leg_kwds)
<AxesSubplot:>
Public Health Centers Data#
base_lima.geometry.iloc[1:2]
#NOTH (x), EAST(Y)
9090 POLYGON ((-77.06517 -11.57512, -77.06505 -11.5...
Name: geometry, dtype: geometry
# import csv file of public health centers: MINSA, ESSALUD and Regional Goverment
# coordinates information (NORTE: longitud) (ESTE: latitud)
health_centers = pd.read_csv(r'../_data/centros_hospitalarios.csv')
health_centers.head( 5 )
| Institución | Código Único | Nombre del establecimiento | Clasificación | Tipo | Departamento | Provincia | Distrito | UBIGEO | Dirección | ... | Estado | Situación | Condición | Inspección | NORTE | ESTE | COTA | CAMAS | code1 | code2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | GOBIERNO REGIONAL | 2076 | SAN PABLO | PUESTOS DE SALUD O POSTAS DE SALUD | ESTABLECIMIENTO DE SALUD SIN INTERNAMIENTO | PIURA | TALARA | LA BREA | 200703 | LAMBAYEQUE S/N NEGRITOS | ... | ACTIVADO | EN FUNCIONAMIENTO | -81.310632 | -4.665043 | 15.3 | NaN | 19 | 1 | ||
| 1 | ESSALUD | 20628 | POSTA MEDICA NEGRITOS | PUESTOS DE SALUD O POSTAS DE SALUD | ESTABLECIMIENTO DE SALUD SIN INTERNAMIENTO | PIURA | TALARA | LA BREA | 200703 | AVENIDA SAN MARTIN Nº 100 NEGRITOS | ... | ACTIVADO | EN FUNCIONAMIENTO | -81.306508 | -4.654384 | 20.4 | NaN | 19 | 1 | ||
| 2 | GOBIERNO REGIONAL | 2075 | NEGRITOS | CENTROS DE SALUD O CENTROS MEDICOS | ESTABLECIMIENTO DE SALUD SIN INTERNAMIENTO | PIURA | TALARA | LA BREA | 200703 | URB. SUD AMERICA D-14 TALARA | ... | ACTIVADO | EN FUNCIONAMIENTO | -81.303974 | -4.658634 | 41.6 | NaN | 4 | 1 | ||
| 3 | GOBIERNO REGIONAL | 2077 | LOBITOS | PUESTOS DE SALUD O POSTAS DE SALUD | ESTABLECIMIENTO DE SALUD SIN INTERNAMIENTO | PIURA | TALARA | LOBITOS | 200704 | BARRIO PRIMAVERA A- 100 LOBITOS | ... | ACTIVADO | EN FUNCIONAMIENTO | -81.275139 | -4.456625 | 39.4 | NaN | 19 | 1 | ||
| 4 | ESSALUD | 12243 | HOSPITAL II ESSALUD TALARA | HOSPITALES O CLINICAS DE ATENCION GENERAL | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | PIURA | TALARA | PARIÑAS | 200701 | AV. PANAMERICANA S/N CARRETERA A LOBITOS | ... | ACTIVADO | EN FUNCIONAMIENTO | -81.273840 | -4.568419 | 28.7 | 35.0 | 12 | 3 |
5 rows × 35 columns
health_centers.columns
Index(['Institución', 'Código Único', 'Nombre del establecimiento',
'Clasificación', 'Tipo', 'Departamento', 'Provincia', 'Distrito',
'UBIGEO', 'Dirección', 'Código DISA', 'Código Red', 'Código Microrred',
'DISA', 'Red', 'Microrred', 'Código UE', 'Unidad Ejecutora',
'Categoria', 'Teléfono', 'Tipo Doc.Categorización',
'Nro.Doc.Categorización', 'Horario', 'Inicio de Actividad',
'Director Médico y/o Responsable de la Atención de Salud', 'Estado',
'Situación', 'Condición', 'Inspección', 'NORTE', 'ESTE', 'COTA',
'CAMAS', 'code1', 'code2'],
dtype='object')
health_centers.NORTE
0 -81.310632
1 -81.306508
2 -81.303974
3 -81.275139
4 -81.273840
...
8459 NaN
8460 NaN
8461 NaN
8462 NaN
8463 NaN
Name: NORTE, Length: 8464, dtype: float64
# DataFrame to GeoDataFrame
Public_health = gpd.GeoDataFrame( health_centers, geometry=gpd.points_from_xy( health_centers.NORTE, health_centers.ESTE ) )
Public_health.head( 5 )
| Institución | Código Único | Nombre del establecimiento | Clasificación | Tipo | Departamento | Provincia | Distrito | UBIGEO | Dirección | ... | Situación | Condición | Inspección | NORTE | ESTE | COTA | CAMAS | code1 | code2 | geometry | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | GOBIERNO REGIONAL | 2076 | SAN PABLO | PUESTOS DE SALUD O POSTAS DE SALUD | ESTABLECIMIENTO DE SALUD SIN INTERNAMIENTO | PIURA | TALARA | LA BREA | 200703 | LAMBAYEQUE S/N NEGRITOS | ... | EN FUNCIONAMIENTO | -81.310632 | -4.665043 | 15.3 | NaN | 19 | 1 | POINT (-81.31063 -4.66504) | ||
| 1 | ESSALUD | 20628 | POSTA MEDICA NEGRITOS | PUESTOS DE SALUD O POSTAS DE SALUD | ESTABLECIMIENTO DE SALUD SIN INTERNAMIENTO | PIURA | TALARA | LA BREA | 200703 | AVENIDA SAN MARTIN Nº 100 NEGRITOS | ... | EN FUNCIONAMIENTO | -81.306508 | -4.654384 | 20.4 | NaN | 19 | 1 | POINT (-81.30651 -4.65438) | ||
| 2 | GOBIERNO REGIONAL | 2075 | NEGRITOS | CENTROS DE SALUD O CENTROS MEDICOS | ESTABLECIMIENTO DE SALUD SIN INTERNAMIENTO | PIURA | TALARA | LA BREA | 200703 | URB. SUD AMERICA D-14 TALARA | ... | EN FUNCIONAMIENTO | -81.303974 | -4.658634 | 41.6 | NaN | 4 | 1 | POINT (-81.30397 -4.65863) | ||
| 3 | GOBIERNO REGIONAL | 2077 | LOBITOS | PUESTOS DE SALUD O POSTAS DE SALUD | ESTABLECIMIENTO DE SALUD SIN INTERNAMIENTO | PIURA | TALARA | LOBITOS | 200704 | BARRIO PRIMAVERA A- 100 LOBITOS | ... | EN FUNCIONAMIENTO | -81.275139 | -4.456625 | 39.4 | NaN | 19 | 1 | POINT (-81.27514 -4.45663) | ||
| 4 | ESSALUD | 12243 | HOSPITAL II ESSALUD TALARA | HOSPITALES O CLINICAS DE ATENCION GENERAL | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | PIURA | TALARA | PARIÑAS | 200701 | AV. PANAMERICANA S/N CARRETERA A LOBITOS | ... | EN FUNCIONAMIENTO | -81.273840 | -4.568419 | 28.7 | 35.0 | 12 | 3 | POINT (-81.27384 -4.56842) |
5 rows × 36 columns
# Setting UTM Zones (South): "EPSG:32733" #depending on the zone, it is PERU
# Public_health = Public_health.set_crs("EPSG:32733")
Public_health = Public_health.set_crs( "EPSG:32718", allow_override = True)
Public_health.crs
<Projected CRS: EPSG:32718>
Name: WGS 84 / UTM zone 18S
Axis Info [cartesian]:
- E[east]: Easting (metre)
- N[north]: Northing (metre)
Area of Use:
- name: World - S hemisphere - 78°W to 72°W - by country
- bounds: (-78.0, -80.0, -72.0, 0.0)
Coordinate Operation:
- name: UTM zone 18S
- method: Transverse Mercator
Datum: World Geodetic System 1984
- Ellipsoid: WGS 84
- Prime Meridian: Greenwich
Public_health.head( 5 )
| Institución | Código Único | Nombre del establecimiento | Clasificación | Tipo | Departamento | Provincia | Distrito | UBIGEO | Dirección | ... | Situación | Condición | Inspección | NORTE | ESTE | COTA | CAMAS | code1 | code2 | geometry | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | GOBIERNO REGIONAL | 2076 | SAN PABLO | PUESTOS DE SALUD O POSTAS DE SALUD | ESTABLECIMIENTO DE SALUD SIN INTERNAMIENTO | PIURA | TALARA | LA BREA | 200703 | LAMBAYEQUE S/N NEGRITOS | ... | EN FUNCIONAMIENTO | -81.310632 | -4.665043 | 15.3 | NaN | 19 | 1 | POINT (-81.311 -4.665) | ||
| 1 | ESSALUD | 20628 | POSTA MEDICA NEGRITOS | PUESTOS DE SALUD O POSTAS DE SALUD | ESTABLECIMIENTO DE SALUD SIN INTERNAMIENTO | PIURA | TALARA | LA BREA | 200703 | AVENIDA SAN MARTIN Nº 100 NEGRITOS | ... | EN FUNCIONAMIENTO | -81.306508 | -4.654384 | 20.4 | NaN | 19 | 1 | POINT (-81.307 -4.654) | ||
| 2 | GOBIERNO REGIONAL | 2075 | NEGRITOS | CENTROS DE SALUD O CENTROS MEDICOS | ESTABLECIMIENTO DE SALUD SIN INTERNAMIENTO | PIURA | TALARA | LA BREA | 200703 | URB. SUD AMERICA D-14 TALARA | ... | EN FUNCIONAMIENTO | -81.303974 | -4.658634 | 41.6 | NaN | 4 | 1 | POINT (-81.304 -4.659) | ||
| 3 | GOBIERNO REGIONAL | 2077 | LOBITOS | PUESTOS DE SALUD O POSTAS DE SALUD | ESTABLECIMIENTO DE SALUD SIN INTERNAMIENTO | PIURA | TALARA | LOBITOS | 200704 | BARRIO PRIMAVERA A- 100 LOBITOS | ... | EN FUNCIONAMIENTO | -81.275139 | -4.456625 | 39.4 | NaN | 19 | 1 | POINT (-81.275 -4.457) | ||
| 4 | ESSALUD | 12243 | HOSPITAL II ESSALUD TALARA | HOSPITALES O CLINICAS DE ATENCION GENERAL | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | PIURA | TALARA | PARIÑAS | 200701 | AV. PANAMERICANA S/N CARRETERA A LOBITOS | ... | EN FUNCIONAMIENTO | -81.273840 | -4.568419 | 28.7 | 35.0 | 12 | 3 | POINT (-81.274 -4.568) |
5 rows × 36 columns
Types and classification public centers#
We focus on Postas médicas and Hospitales#
code2 (Type)#
ESTABLECIMIENTO DE SALUD SIN INTERNAMIENTO: 1
SERVICIO MÉDICO DE APOYO: 2
ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO: 3
code1 (Clasification)#
ATENCION PRE HOSPITALARIA: 1
CENTROS DE ATENCION PARA DEPENDIENTES A SUSTANCIAS PSICOACTIVAS Y OTRAS DEPENDENCIAS: 2
CENTROS DE SALUD CON CAMAS DE INTERNAMIENTO,CENTROS DE SALUD CON CAMAS DE INTERNAMIENTO: 3
CENTROS DE SALUD O CENTROS MEDICOS: 4
CENTROS DE SALUD O CENTROS MEDICOS,CENTROS DE SALUD O CENTROS MEDICOS: 5
CENTROS DE VACUNACION: 6
CENTROS MEDICOS ESPECIALIZADOS: 7
CONSULTORIOS MEDICOS Y DE OTROS PROFESIONALES DE LA SALUD: 8
ESTABLECIMIENTOS DE RECUPERACION O REPOSO: 9
HEMODIALISIS: 10
HOSPITALES O CLINICAS DE ATENCION ESPECIALIZADA: 11
HOSPITALES O CLINICAS DE ATENCION GENERAL: 12
INSTITUTOS DE SALUD ESPECIALIZADOS: 13
MEDICINA FISICA,REHABILITACION: 14
PATOLOGIA CLINICA: 15
PATOLOGIA CLINICA,ANATOMIA PATOLOGICA: 16
PATOLOGIA CLINICA,DIAGNOSTICO POR IMAGENES: 17
POLICLINICOS: 18
PUESTOS DE SALUD O POSTAS DE SALUD: 19
PUESTOS DE SALUD O POSTAS DE SALUD,PUESTOS DE SALUD O POSTAS DE SALUD: 19
SERVICIO DE TRASLADO DE PACIENTES: 20
SERVICIO DE TRASLADO DE PACIENTES,ATENCION DOMICILIARIA: 21
SERVICIO DE TRASLADO DE PACIENTES,ATENCION DOMICILIARIA,ATENCION PRE HOSPITALARIA: 22
SERVICIO DE TRASLADO DE PACIENTES,ATENCION PRE HOSPITALARIA: 23
CENTROS DE SALUD CON CAMAS DE INTERNAMIENTO: 24
Public_health.head( 5 )
| Institución | Código Único | Nombre del establecimiento | Clasificación | Tipo | Departamento | Provincia | Distrito | UBIGEO | Dirección | ... | Situación | Condición | Inspección | NORTE | ESTE | COTA | CAMAS | code1 | code2 | geometry | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | GOBIERNO REGIONAL | 2076 | SAN PABLO | PUESTOS DE SALUD O POSTAS DE SALUD | ESTABLECIMIENTO DE SALUD SIN INTERNAMIENTO | PIURA | TALARA | LA BREA | 200703 | LAMBAYEQUE S/N NEGRITOS | ... | EN FUNCIONAMIENTO | -81.310632 | -4.665043 | 15.3 | NaN | 19 | 1 | POINT (-81.311 -4.665) | ||
| 1 | ESSALUD | 20628 | POSTA MEDICA NEGRITOS | PUESTOS DE SALUD O POSTAS DE SALUD | ESTABLECIMIENTO DE SALUD SIN INTERNAMIENTO | PIURA | TALARA | LA BREA | 200703 | AVENIDA SAN MARTIN Nº 100 NEGRITOS | ... | EN FUNCIONAMIENTO | -81.306508 | -4.654384 | 20.4 | NaN | 19 | 1 | POINT (-81.307 -4.654) | ||
| 2 | GOBIERNO REGIONAL | 2075 | NEGRITOS | CENTROS DE SALUD O CENTROS MEDICOS | ESTABLECIMIENTO DE SALUD SIN INTERNAMIENTO | PIURA | TALARA | LA BREA | 200703 | URB. SUD AMERICA D-14 TALARA | ... | EN FUNCIONAMIENTO | -81.303974 | -4.658634 | 41.6 | NaN | 4 | 1 | POINT (-81.304 -4.659) | ||
| 3 | GOBIERNO REGIONAL | 2077 | LOBITOS | PUESTOS DE SALUD O POSTAS DE SALUD | ESTABLECIMIENTO DE SALUD SIN INTERNAMIENTO | PIURA | TALARA | LOBITOS | 200704 | BARRIO PRIMAVERA A- 100 LOBITOS | ... | EN FUNCIONAMIENTO | -81.275139 | -4.456625 | 39.4 | NaN | 19 | 1 | POINT (-81.275 -4.457) | ||
| 4 | ESSALUD | 12243 | HOSPITAL II ESSALUD TALARA | HOSPITALES O CLINICAS DE ATENCION GENERAL | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | PIURA | TALARA | PARIÑAS | 200701 | AV. PANAMERICANA S/N CARRETERA A LOBITOS | ... | EN FUNCIONAMIENTO | -81.273840 | -4.568419 | 28.7 | 35.0 | 12 | 3 | POINT (-81.274 -4.568) |
5 rows × 36 columns
# code1 = 19 to postas médicas
# code1 = 12 & 24 to Hospitals
Public_health1 = Public_health[Public_health.code1 == "19"]
Public_health2 = Public_health[(Public_health.code1 == "12") | (Public_health.code1 == "24")]
Public_health2.head( 5 )
| Institución | Código Único | Nombre del establecimiento | Clasificación | Tipo | Departamento | Provincia | Distrito | UBIGEO | Dirección | ... | Situación | Condición | Inspección | NORTE | ESTE | COTA | CAMAS | code1 | code2 | geometry | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | ESSALUD | 12243 | HOSPITAL II ESSALUD TALARA | HOSPITALES O CLINICAS DE ATENCION GENERAL | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | PIURA | TALARA | PARIÑAS | 200701 | AV. PANAMERICANA S/N CARRETERA A LOBITOS | ... | EN FUNCIONAMIENTO | -81.273840 | -4.568419 | 28.7 | 35.0 | 12 | 3 | POINT (-81.274 -4.568) | ||
| 5 | GOBIERNO REGIONAL | 2074 | TALARA II | CENTROS DE SALUD CON CAMAS DE INTERNAMIENTO | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | PIURA | TALARA | PARIÑAS | 200701 | AVENIDA GRAU S/N EXPOLICLINICO PETROPERU | ... | EN FUNCIONAMIENTO | -81.272096 | -4.578829 | 21.1 | NaN | 24 | 3 | POINT (-81.272 -4.579) | ||
| 19 | ESSALUD | 11763 | HOSPITAL I MIGUEL CRUZADO VERA - PAITA | CENTROS DE SALUD CON CAMAS DE INTERNAMIENTO | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | PIURA | PAITA | PAITA | 200501 | CARRETERA PAITA - PIURA KM 1.5 | ... | EN FUNCIONAMIENTO | -81.100243 | -5.087583 | 75.9 | 12.0 | 24 | 3 | POINT (-81.100 -5.088) | ||
| 20 | GOBIERNO REGIONAL | 2021 | HOSPITAL LAS MERCEDES-PAITA | HOSPITALES O CLINICAS DE ATENCION GENERAL | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | PIURA | PAITA | PAITA | 200501 | LOS PESCADORES S/N LA PUNTA | ... | EN FUNCIONAMIENTO | -81.091531 | -5.090815 | 78.3 | 79.0 | 12 | 3 | POINT (-81.092 -5.091) | ||
| 23 | GOBIERNO REGIONAL | 2026 | PUEBLO NUEVO DE COLAN | CENTROS DE SALUD CON CAMAS DE INTERNAMIENTO | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | PIURA | PAITA | COLAN | 200504 | JR. ALFONSO UGARTE S/N | ... | EN FUNCIONAMIENTO | -81.058005 | -4.911551 | 16.6 | NaN | 24 | 3 | POINT (-81.058 -4.912) |
5 rows × 36 columns
# # Quantiles quantity of infection
# base_lima.plot(column='Casos', cmap='Set2', figsize=(20, 20), linestyle=':',
# edgecolor='black',
# legend = True)
# Postas Médicas and Hospitales públicos - LIMA
PH_lima_postas = Public_health1[Public_health1.Provincia == "LIMA"]
PH_lima_hospitales = Public_health2[Public_health2.Provincia == "LIMA"]
PH_lima_hospitales
| Institución | Código Único | Nombre del establecimiento | Clasificación | Tipo | Departamento | Provincia | Distrito | UBIGEO | Dirección | ... | Situación | Condición | Inspección | NORTE | ESTE | COTA | CAMAS | code1 | code2 | geometry | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2715 | MINSA | 5814 | CENTRO DE SALUD MATERNO INFANTIL ANCON | CENTROS DE SALUD CON CAMAS DE INTERNAMIENTO | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | ANCON | 150102 | JR. LOA 595 ANCON | ... | EN FUNCIONAMIENTO | -77.172725 | -11.774719 | 12.0 | NaN | 24 | 3 | POINT (-77.173 -11.775) | ||
| 2803 | MINSA | 5816 | MATERNO INFANTIL DR. ENRIQUE MARTIN ALTUNA | CENTROS DE SALUD CON CAMAS DE INTERNAMIENTO | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | PUENTE PIEDRA | 150125 | ASOC VIV E INDUSTRIAS VIRGEN DE LAS NIEVES URB... | ... | EN FUNCIONAMIENTO | -77.109100 | -11.837611 | 200.0 | NaN | 24 | 3 | POINT (-77.109 -11.838) | ||
| 2832 | ESSALUD | 8564 | HOSPITAL I OCTAVIO MONGRUT MUÑOZ | HOSPITALES O CLINICAS DE ATENCION GENERAL | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | SAN MIGUEL | 150136 | AV PARQUE DE LAS LEYENDAS 255 | ... | EN FUNCIONAMIENTO | -77.094583 | -12.065961 | NaN | 49.0 | 12 | 3 | POINT (-77.095 -12.066) | ||
| 2852 | MINSA | 5817 | SANTA ROSA | CENTROS DE SALUD CON CAMAS DE INTERNAMIENTO | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | PUENTE PIEDRA | 150125 | AA.HH. SANTA ROSA, MZ. 50, LOTE PM - AV. SANTA... | ... | EN FUNCIONAMIENTO | -77.081693 | -11.874827 | 200.0 | NaN | 24 | 3 | POINT (-77.082 -11.875) | ||
| 2859 | MINSA | 7632 | HOSPITAL CARLOS LANFRANCO LA HOZ | HOSPITALES O CLINICAS DE ATENCION GENERAL | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | PUENTE PIEDRA | 150125 | AV. SAENZ PEÑA - CUADRA 6 S/N | ... | EN FUNCIONAMIENTO | -77.079335 | -11.863181 | NaN | 102.0 | 12 | 3 | POINT (-77.079 -11.863) | ||
| 2862 | MINSA | 5799 | CLAS JUAN PABLO II | CENTROS DE SALUD CON CAMAS DE INTERNAMIENTO | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | LOS OLIVOS | 150117 | CALLE 28 S/N AA.HH. JUAN PABLO II | ... | EN FUNCIONAMIENTO | -77.078891 | -11.953212 | 63.0 | NaN | 24 | 3 | POINT (-77.079 -11.953) | ||
| 2875 | MINSA | 5821 | LOS SUREÑOS | CENTROS DE SALUD CON CAMAS DE INTERNAMIENTO | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | PUENTE PIEDRA | 150125 | COOP. VIVIENDA LOS SUREÑOS MZ M LT. 35 | ... | EN FUNCIONAMIENTO | -77.069701 | -11.887532 | 200.0 | NaN | 24 | 3 | POINT (-77.070 -11.888) | ||
| 2879 | MINSA | 6196 | MAGDALENA | CENTROS DE SALUD CON CAMAS DE INTERNAMIENTO | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | MAGDALENA DEL MAR | 150120 | JR. JUNÍN 322 | ... | EN FUNCIONAMIENTO | -77.068627 | -12.088694 | 27.0 | NaN | 24 | 3 | POINT (-77.069 -12.089) | ||
| 2887 | MINSA | 6214 | HOSPITAL VICTOR LARCO HERRERA | HOSPITALES O CLINICAS DE ATENCION GENERAL | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | MAGDALENA DEL MAR | 150120 | AV.AUGUSTO PEREZ ARANIBAR N° 600 | ... | EN FUNCIONAMIENTO | -77.065756 | -12.098705 | NaN | 590.0 | 12 | 3 | POINT (-77.066 -12.099) | ||
| 2904 | MINSA | 5772 | CENTRO MATERNO INFANTIL SANTA LUZMILA II | CENTROS DE SALUD CON CAMAS DE INTERNAMIENTO | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | COMAS | 150110 | AVENIDA AV. 22 DE AGOSTO N° 1001- URB SANTA LU... | ... | EN FUNCIONAMIENTO | -77.058712 | -11.947025 | 101.0 | NaN | 24 | 3 | POINT (-77.059 -11.947) | ||
| 2905 | ESSALUD | 8671 | HOSPITAL I MARINO MOLINA SCIPPA | HOSPITALES O CLINICAS DE ATENCION GENERAL | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | COMAS | 150110 | AV. GUILLERMO DE LA FUENTE NO 515 URB. SANTA L... | ... | EN FUNCIONAMIENTO | -77.058242 | -11.943809 | NaN | 106.0 | 12 | 3 | POINT (-77.058 -11.944) | ||
| 2912 | MINSA | 5755 | CENTRO MATERNO INFANTIL LAURA RODRIGUEZ | CENTROS DE SALUD CON CAMAS DE INTERNAMIENTO | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | COMAS | 150110 | CALLE CALLE 30 N° 150 (ANTES MZ R1 LOTE 2- PAR... | ... | EN FUNCIONAMIENTO | -77.055362 | -11.916050 | 101.0 | NaN | 24 | 3 | POINT (-77.055 -11.916) | ||
| 2927 | MINSA | 5791 | TAHUANTINSUYO BAJO | CENTROS DE SALUD CON CAMAS DE INTERNAMIENTO | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | INDEPENDENCIA | 150112 | AVENIDA AV. CHINCHAYSUYO CUADRA 4 - URB. TAHUA... | ... | EN FUNCIONAMIENTO | -77.052983 | -11.978466 | 85.0 | NaN | 24 | 3 | POINT (-77.053 -11.978) | ||
| 2943 | ESSALUD | 9188 | HOSPITAL II RAMÓN CASTILLA | HOSPITALES O CLINICAS DE ATENCION GENERAL | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | LIMA | 150101 | JIRÓN JIRÓN GUILLERMO DANSEY Nº 390 JIRÓN GUIL... | ... | EN FUNCIONAMIENTO | -77.044033 | -12.044341 | NaN | 49.0 | 12 | 3 | POINT (-77.044 -12.044) | ||
| 2955 | MINSA | 6207 | NACIONAL ARZOBISPO LOAYZA | HOSPITALES O CLINICAS DE ATENCION GENERAL | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | LIMA | 150101 | AV. ALFONSO UGARTE 848 | ... | EN FUNCIONAMIENTO | -77.042560 | -12.049845 | NaN | 662.0 | 12 | 3 | POINT (-77.043 -12.050) | ||
| 2984 | MINSA | 5644 | RIMAC | CENTROS DE SALUD CON CAMAS DE INTERNAMIENTO | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | RIMAC | 150128 | PASAJE SAN GERMAN Nº 270 URB. VILLACAMPA – RIMAC | ... | EN FUNCIONAMIENTO | -77.033661 | -12.034340 | NaN | NaN | 24 | 3 | POINT (-77.034 -12.034) | ||
| 2988 | ESSALUD | 8502 | HOSPITAL III EMERGENCIAS GRAU | HOSPITALES O CLINICAS DE ATENCION GENERAL | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | LIMA | 150101 | AVENIDA GRAU N° 351 | ... | EN FUNCIONAMIENTO | -77.031278 | -12.058957 | NaN | 231.0 | 12 | 3 | POINT (-77.031 -12.059) | ||
| 2997 | ESSALUD | 8489 | ESSALUD - HOSPITAL II SUÁREZ ANGAMOS | HOSPITALES O CLINICAS DE ATENCION GENERAL | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | MIRAFLORES | 150122 | AV. ANGAMOS ESTE Nº 261 | ... | EN FUNCIONAMIENTO | -77.027993 | -12.113623 | NaN | 84.0 | 12 | 3 | POINT (-77.028 -12.114) | ||
| 3009 | MINSA | 6162 | CENTRO DE EMERGENCIAS SAN PEDRO DE LOS CHORRILLOS | CENTROS DE SALUD CON CAMAS DE INTERNAMIENTO | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | CHORRILLOS | 150108 | CALLE FERROCARRIL S/N | ... | EN FUNCIONAMIENTO | -77.023590 | -12.166438 | 46.0 | NaN | 24 | 3 | POINT (-77.024 -12.166) | ||
| 3017 | MINSA | 6178 | CENTRO DE SALUD SURQUILLO | CENTROS DE SALUD CON CAMAS DE INTERNAMIENTO | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | SURQUILLO | 150141 | JR. NARCISO DE LA COLINA 840 | ... | EN FUNCIONAMIENTO | -77.022067 | -12.118238 | 122.0 | NaN | 24 | 3 | POINT (-77.022 -12.118) | ||
| 3023 | MINSA | 6175 | EL PORVENIR | CENTROS DE SALUD CON CAMAS DE INTERNAMIENTO | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | LA VICTORIA | 150115 | JR. SEBASTIÁN BARRANCA 977 | ... | EN FUNCIONAMIENTO | -77.020907 | -12.067660 | 142.0 | NaN | 24 | 3 | POINT (-77.021 -12.068) | ||
| 3027 | MINSA | 5991 | CENTRO MATERNO INFANTIL DE SALUD - "VIRGEN DEL... | CENTROS DE SALUD CON CAMAS DE INTERNAMIENTO | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | CHORRILLOS | 150108 | CALLE LEOPOLDO ARIAS NRO. 200 | ... | EN FUNCIONAMIENTO | -77.020298 | -12.165916 | 46.0 | NaN | 24 | 3 | POINT (-77.020 -12.166) | ||
| 3036 | MINSA | 5735 | EL PROGRESO | CENTROS DE SALUD CON CAMAS DE INTERNAMIENTO | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | CARABAYLLO | 150106 | AVENIDA AV. TUPAC AMARU Nº 2950 (KM 22) DISTRI... | ... | EN FUNCIONAMIENTO | -77.016728 | -11.875540 | 221.0 | NaN | 24 | 3 | POINT (-77.017 -11.876) | ||
| 3040 | MINSA | 6206 | NACIONAL DOS DE MAYO | HOSPITALES O CLINICAS DE ATENCION GENERAL | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | LIMA | 150101 | PARQUE HISTORIA DE LA MEDICINA PERUANA S/N ALT... | ... | EN FUNCIONAMIENTO | -77.016269 | -12.057170 | NaN | 594.0 | 12 | 3 | POINT (-77.016 -12.057) | ||
| 3048 | MINSA | 5842 | PIEDRA LIZA | CENTROS DE SALUD CON CAMAS DE INTERNAMIENTO | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | RIMAC | 150128 | AV. SANTA ROSA S/N - BARRIO FISCAL PIEDRA LIZA | ... | EN FUNCIONAMIENTO | -77.012911 | -12.032531 | 205.0 | NaN | 24 | 3 | POINT (-77.013 -12.033) | ||
| 3071 | MINSA | 5998 | BUENOS AIRES DE VILLA | CENTROS DE SALUD CON CAMAS DE INTERNAMIENTO | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | CHORRILLOS | 150108 | AV. BUENOS AIRES DE VILLA S/N. | ... | EN FUNCIONAMIENTO | -77.004874 | -12.188766 | 46.0 | NaN | 24 | 3 | POINT (-77.005 -12.189) | ||
| 3077 | MINSA | 5617 | HOSPITAL SAN JUAN DE LURIGANCHO | HOSPITALES O CLINICAS DE ATENCION GENERAL | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | SAN JUAN DE LURIGANCHO | 150132 | ENTRE PARADERO 10 Y 11 DE AV. CANTO GRANDE (HU... | ... | EN FUNCIONAMIENTO | -77.003057 | -11.966577 | NaN | 150.0 | 12 | 3 | POINT (-77.003 -11.967) | ||
| 3093 | MINSA | 5966 | SAN FERNANDO | CENTROS DE SALUD CON CAMAS DE INTERNAMIENTO | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | ATE | 150103 | JIRÓNJOSE SANTOS CHOCANO CDRA.01 S/N - URB.VAL... | ... | EN FUNCIONAMIENTO | -76.996408 | -12.060061 | 349.0 | 6.0 | 24 | 3 | POINT (-76.996 -12.060) | ||
| 3096 | ESSALUD | 8317 | HOSPITAL AURELIO DIAZ UFANO Y PERAL | HOSPITALES O CLINICAS DE ATENCION GENERAL | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | SAN JUAN DE LURIGANCHO | 150132 | CALLE MAJES S/N G-11 - URB. LOS PINOS | ... | EN FUNCIONAMIENTO | -76.995567 | -11.968478 | NaN | 54.0 | 12 | 3 | POINT (-76.996 -11.968) | ||
| 3161 | MINSA | 6750 | SANTA ANITA | CENTROS DE SALUD CON CAMAS DE INTERNAMIENTO | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | SANTA ANITA | 150137 | CALLE LOS MOCHICAS S/N PARQUE LAMPA DE ORO CO... | ... | EN FUNCIONAMIENTO | -76.969171 | -12.041362 | NaN | 6.0 | 24 | 3 | POINT (-76.969 -12.041) | ||
| 3164 | MINSA | 6104 | MANUEL BARRETO | CENTROS DE SALUD CON CAMAS DE INTERNAMIENTO | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | SAN JUAN DE MIRAFLORES | 150133 | JR. MANUEL BARRETO S/N ZONA K-CIUDAD DE DIOS | ... | EN FUNCIONAMIENTO | -76.968442 | -12.152256 | 108.0 | NaN | 24 | 3 | POINT (-76.968 -12.152) | ||
| 3175 | MINSA | 6107 | OLLANTAY | CENTROS DE SALUD CON CAMAS DE INTERNAMIENTO | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | SAN JUAN DE MIRAFLORES | 150133 | AV. PROLONGACION GABRIEL TORRES S/N. PAMPLONA ... | ... | EN FUNCIONAMIENTO | -76.963548 | -12.135808 | 108.0 | NaN | 24 | 3 | POINT (-76.964 -12.136) | ||
| 3204 | MINSA | 6152 | JOSE CARLOS MARIATEGUI | CENTROS DE SALUD CON CAMAS DE INTERNAMIENTO | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | VILLA MARIA DEL TRIUNFO | 150143 | AV. PRIMAVERA CUADRA 23 S/N JOSE CARLOS MARIAT... | ... | EN FUNCIONAMIENTO | -76.950802 | -12.147521 | 163.0 | NaN | 24 | 3 | POINT (-76.951 -12.148) | ||
| 3213 | MINSA | 6132 | CENTRO MATERNO INFANTIL SAN JOSE | CENTROS DE SALUD CON CAMAS DE INTERNAMIENTO | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | VILLA EL SALVADOR | 150142 | AV. LOS ÁNGELES S/N, SECTOR 1, GRUPO 15 | ... | EN FUNCIONAMIENTO | -76.946310 | -12.198297 | 143.0 | NaN | 24 | 3 | POINT (-76.946 -12.198) | ||
| 3214 | ESSALUD | 10135 | HOSPITAL I CARLOS ALCANTARA BUTTERFIELD | HOSPITALES O CLINICAS DE ATENCION GENERAL | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | LA MOLINA | 150114 | AV. LOS CONSTRUCTORES 1201 - URB. COVIMA | ... | EN FUNCIONAMIENTO | -76.945544 | -12.062131 | NaN | 50.0 | 12 | 3 | POINT (-76.946 -12.062) | ||
| 3216 | MINSA | 6153 | DANIEL ALCIDES CARRION | CENTROS DE SALUD CON CAMAS DE INTERNAMIENTO | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | VILLA MARIA DEL TRIUNFO | 150143 | AV. PACHACUTEC Nº 3470 | ... | EN FUNCIONAMIENTO | -76.945390 | -12.177699 | 163.0 | NaN | 24 | 3 | POINT (-76.945 -12.178) | ||
| 3217 | MINSA | 6133 | CENTRO MATERNO INFANTIL JUAN PABLO II | CENTROS DE SALUD CON CAMAS DE INTERNAMIENTO | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | VILLA EL SALVADOR | 150142 | AV. MARIANO PASTOR SEVILLA S/N ,SECTOR 6,GRUPO 6 | ... | EN FUNCIONAMIENTO | -76.945290 | -12.219161 | 143.0 | NaN | 24 | 3 | POINT (-76.945 -12.219) | ||
| 3230 | ESSALUD | 18580 | GUILLERMO KAELIN DE LA FUENTE | HOSPITALES O CLINICAS DE ATENCION GENERAL | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | VILLA MARIA DEL TRIUNFO | 150143 | AVENIDA PP.JJ. NUEVA ESPERANZA MZ 1B SUBLOTE 1... | ... | EN FUNCIONAMIENTO | -76.940599 | -12.178289 | NaN | 199.0 | 12 | 3 | POINT (-76.941 -12.178) | ||
| 3238 | MINSA | 6151 | VILLA MARIA DEL TRIUNFO | CENTROS DE SALUD CON CAMAS DE INTERNAMIENTO | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | VILLA MARIA DEL TRIUNFO | 150143 | AV. PEDRO VALLE S/N (ALTURA CUADRA 18 AV.EL TR... | ... | EN FUNCIONAMIENTO | -76.936502 | -12.156910 | 163.0 | NaN | 24 | 3 | POINT (-76.937 -12.157) | ||
| 3255 | MINSA | 6164 | TABLADA DE LURIN | CENTROS DE SALUD CON CAMAS DE INTERNAMIENTO | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | VILLA MARIA DEL TRIUNFO | 150143 | AV. REPUBLICA Y BILLINGURST S/N 2DO SECTOR | ... | EN FUNCIONAMIENTO | -76.927452 | -12.191033 | 163.0 | NaN | 24 | 3 | POINT (-76.927 -12.191) | ||
| 3262 | MINSA | 6124 | CESAR LOPEZ SILVA | CENTROS DE SALUD CON CAMAS DE INTERNAMIENTO | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | VILLA EL SALVADOR | 150142 | SECTOR IV MZ B1 LT S/N 1RA. ETAPA URB.PACHACAM... | ... | EN FUNCIONAMIENTO | -76.923745 | -12.230178 | 143.0 | NaN | 24 | 3 | POINT (-76.924 -12.230) | ||
| 3269 | MINSA | 5945 | HOSPITAL VITARTE | HOSPITALES O CLINICAS DE ATENCION GENERAL | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | ATE | 150103 | AVENIDA NICOLAS AYLLON 5880-CARRETERA CENTRAL ... | ... | EN FUNCIONAMIENTO | -76.919904 | -12.026042 | NaN | 111.0 | 12 | 3 | POINT (-76.920 -12.026) | ||
| 3284 | MINSA | 6141 | JOSE GALVEZ | CENTROS DE SALUD CON CAMAS DE INTERNAMIENTO | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | VILLA MARIA DEL TRIUNFO | 150143 | CRUCE CALLE ARICA CON AGRICULTURA S/N | ... | EN FUNCIONAMIENTO | -76.907215 | -12.210562 | 163.0 | NaN | 24 | 3 | POINT (-76.907 -12.211) | ||
| 3322 | MINSA | 6079 | LURIN | CENTROS DE SALUD CON CAMAS DE INTERNAMIENTO | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | LURIN | 150119 | JIRÓN GRAU Nº 370 JR. GRAU Nº 370 VILLA EL SAL... | ... | EN FUNCIONAMIENTO | -76.869575 | -12.273670 | 10.0 | NaN | 24 | 3 | POINT (-76.870 -12.274) | ||
| 3363 | MINSA | 5883 | HOSPITAL DE BAJA COMPLEJIDAD HUAYCAN | HOSPITALES O CLINICAS DE ATENCION GENERAL | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | ATE | 150103 | AV JOSÉ C.MARIÁTEGUI S/N ZONA B - A.H.HUAYCÁN | ... | EN FUNCIONAMIENTO | -76.820118 | -12.015889 | NaN | 51.0 | 12 | 3 | POINT (-76.820 -12.016) | ||
| 3370 | MINSA | 5978 | TAMBO VIEJO | CENTROS DE SALUD CON CAMAS DE INTERNAMIENTO | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | CIENEGUILLA | 150109 | AVENIDA AV. MANCO CAPAC S/N - ZONA A - TAMBO V... | ... | EN FUNCIONAMIENTO | -76.818267 | -12.111047 | 267.0 | NaN | 24 | 3 | POINT (-76.818 -12.111) | ||
| 3382 | MINSA | 5938 | MIGUEL GRAU | CENTROS DE SALUD CON CAMAS DE INTERNAMIENTO | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | CHACLACAYO | 150107 | CARRETERA CARRETERA CENTRAL KILOMETRO 19.5 - C... | ... | EN FUNCIONAMIENTO | -76.813638 | -11.987051 | 676.0 | 10.0 | 24 | 3 | POINT (-76.814 -11.987) | ||
| 3508 | MINSA | 5947 | HOSPITAL DE MEDIANA COMPLEJIDAD JOSE AGURTO TELLO | HOSPITALES O CLINICAS DE ATENCION GENERAL | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | LURIGANCHO | 150118 | JR. AREQUIPA 214-218 | ... | EN FUNCIONAMIENTO | -76.693699 | -11.934447 | NaN | 67.0 | 12 | 3 | POINT (-76.694 -11.934) | ||
| 7909 | ESSALUD | 8318 | SEGURO SOCIAL DE SALUD | HOSPITALES O CLINICAS DE ATENCION GENERAL | ESTABLECIMIENTO DE SALUD CON INTERNAMIENTO | LIMA | LIMA | SANTA ANITA | 150137 | CARRETERA CENTRAL KM. 3.5 Nº 1351 | ... | EN FUNCIONAMIENTO | NaN | NaN | NaN | 57.0 | 12 | 3 | POINT (nan nan) |
49 rows × 36 columns
Covid y Postas#
base_lima["Muertes Covid"].hist(bins=30)
<AxesSubplot:>
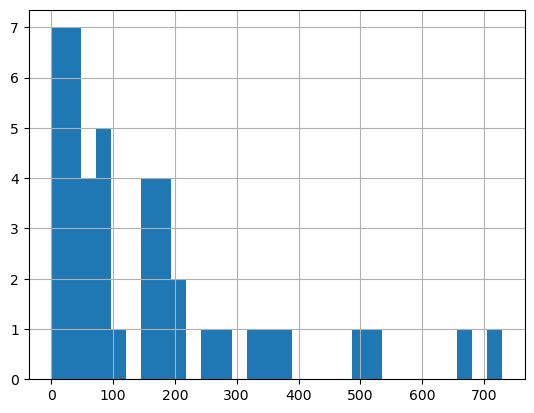
PH_lima_postas
| Institución | Código Único | Nombre del establecimiento | Clasificación | Tipo | Departamento | Provincia | Distrito | UBIGEO | Dirección | ... | Situación | Condición | Inspección | NORTE | ESTE | COTA | CAMAS | code1 | code2 | geometry | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2725 | MINSA | 5824 | SAN JOSE | PUESTOS DE SALUD O POSTAS DE SALUD | ESTABLECIMIENTO DE SALUD SIN INTERNAMIENTO | LIMA | LIMA | ANCON | 150102 | ESQ. CALLE Nº 17 Y CALLE Nº 9 URB. DE INTERES ... | ... | EN FUNCIONAMIENTO | -77.162575 | -11.778237 | 12.0 | NaN | 19 | 1 | POINT (-77.163 -11.778) | ||
| 2728 | MINSA | 14727 | PROFAM | PUESTOS DE SALUD O POSTAS DE SALUD | ESTABLECIMIENTO DE SALUD SIN INTERNAMIENTO | LIMA | LIMA | SANTA ROSA | 150139 | AV. SANTA ROSA, MANZANA O-2 / ASOCIACION DE VI... | ... | EN FUNCIONAMIENTO | -77.159128 | -11.817327 | NaN | NaN | 19 | 1 | POINT (-77.159 -11.817) | ||
| 2740 | MINSA | 16525 | VILLAS DE ANCON | PUESTOS DE SALUD O POSTAS DE SALUD | ESTABLECIMIENTO DE SALUD SIN INTERNAMIENTO | LIMA | LIMA | ANCON | 150102 | MZ X ASOCIACION PRO-VIVIENDA VILLAS DE ANCON | ... | EN FUNCIONAMIENTO | -77.145966 | -11.733970 | 0.0 | NaN | 19 | 1 | POINT (-77.146 -11.734) | ||
| 2762 | MINSA | 5825 | VIRGEN DE LAS MERCEDES | PUESTOS DE SALUD O POSTAS DE SALUD | ESTABLECIMIENTO DE SALUD SIN INTERNAMIENTO | LIMA | LIMA | SANTA ROSA | 150139 | ASOC. VIV. SANTA ROSA MZ. B LT. 09 | ... | EN FUNCIONAMIENTO | -77.131677 | -11.815250 | 12.0 | NaN | 19 | 1 | POINT (-77.132 -11.815) | ||
| 2797 | MINSA | 5750 | CERRO LA REGLA | PUESTOS DE SALUD O POSTAS DE SALUD | ESTABLECIMIENTO DE SALUD SIN INTERNAMIENTO | LIMA | LIMA | SAN MARTIN DE PORRES | 150135 | MZ B LT 11 - AAHH ESPERANZA, COMITÉ 2 | ... | EN FUNCIONAMIENTO | -77.115765 | -11.989613 | 123.0 | NaN | 19 | 1 | POINT (-77.116 -11.990) | ||
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 3486 | MINSA | 5868 | SEÑOR DE LOS MILAGROS | PUESTOS DE SALUD O POSTAS DE SALUD | ESTABLECIMIENTO DE SALUD SIN INTERNAMIENTO | LIMA | LIMA | LURIGANCHO | 150118 | AVENIDA AV.PRECURSORES S/N AAHH NICOLAS DE PIE... | ... | EN FUNCIONAMIENTO | -76.714793 | -11.935857 | 911.0 | NaN | 19 | 1 | POINT (-76.715 -11.936) | ||
| 3516 | MINSA | 5870 | PABLO PATRON | PUESTOS DE SALUD O POSTAS DE SALUD | ESTABLECIMIENTO DE SALUD SIN INTERNAMIENTO | LIMA | LIMA | LURIGANCHO | 150118 | AAHH PABLO PATRON - CALLE 14 MZ. V LOTE 7 DIST... | ... | EN FUNCIONAMIENTO | -76.686205 | -11.930408 | 911.0 | NaN | 19 | 1 | POINT (-76.686 -11.930) | ||
| 3519 | MINSA | 5867 | MARISCAL CASTILLA | PUESTOS DE SALUD O POSTAS DE SALUD | ESTABLECIMIENTO DE SALUD SIN INTERNAMIENTO | LIMA | LIMA | LURIGANCHO | 150118 | CALLE SANTA ROSA MZ O S/N - AAHH MARISCAL CAST... | ... | EN FUNCIONAMIENTO | -76.685491 | -11.933911 | 911.0 | NaN | 19 | 1 | POINT (-76.685 -11.934) | ||
| 8125 | MINSA | 5940 | HUASCATA | PUESTOS DE SALUD O POSTAS DE SALUD | ESTABLECIMIENTO DE SALUD SIN INTERNAMIENTO | LIMA | LIMA | CHACLACAYO | 150107 | OTROS MZ N LOTE 05 AAHH CERRO VECINO HUASCATA-... | ... | EN FUNCIONAMIENTO | NaN | NaN | NaN | NaN | 19 | 1 | POINT (nan nan) | ||
| 8184 | MINSA | 23635 | RED DE SERVICIOS DE SALUD SAN JUAN DE MIRAFLOR... | PUESTOS DE SALUD O POSTAS DE SALUD | ESTABLECIMIENTO DE SALUD SIN INTERNAMIENTO | LIMA | LIMA | LIMA | 150101 | OTROS AA.HH. Virgen del Guadalupe MANZANA E LO... | ... | EN FUNCIONAMIENTO | NaN | NaN | NaN | NaN | 19 | 1 | POINT (nan nan) |
164 rows × 36 columns
# Covid deaths and Postas médicas
fig, ax = plt.subplots(figsize=(20, 20))
#Heatmap Lima and Covid infection
base_lima.plot( column='Muertes Covid',
cmap='Reds',
linestyle='--',
edgecolor='black',
legend = True,
ax=ax )
# Plot in the shapefile (R style)
PH_lima_postas.plot(ax = ax, color = 'blue', markersize=10, edgecolor='black')
<AxesSubplot:>
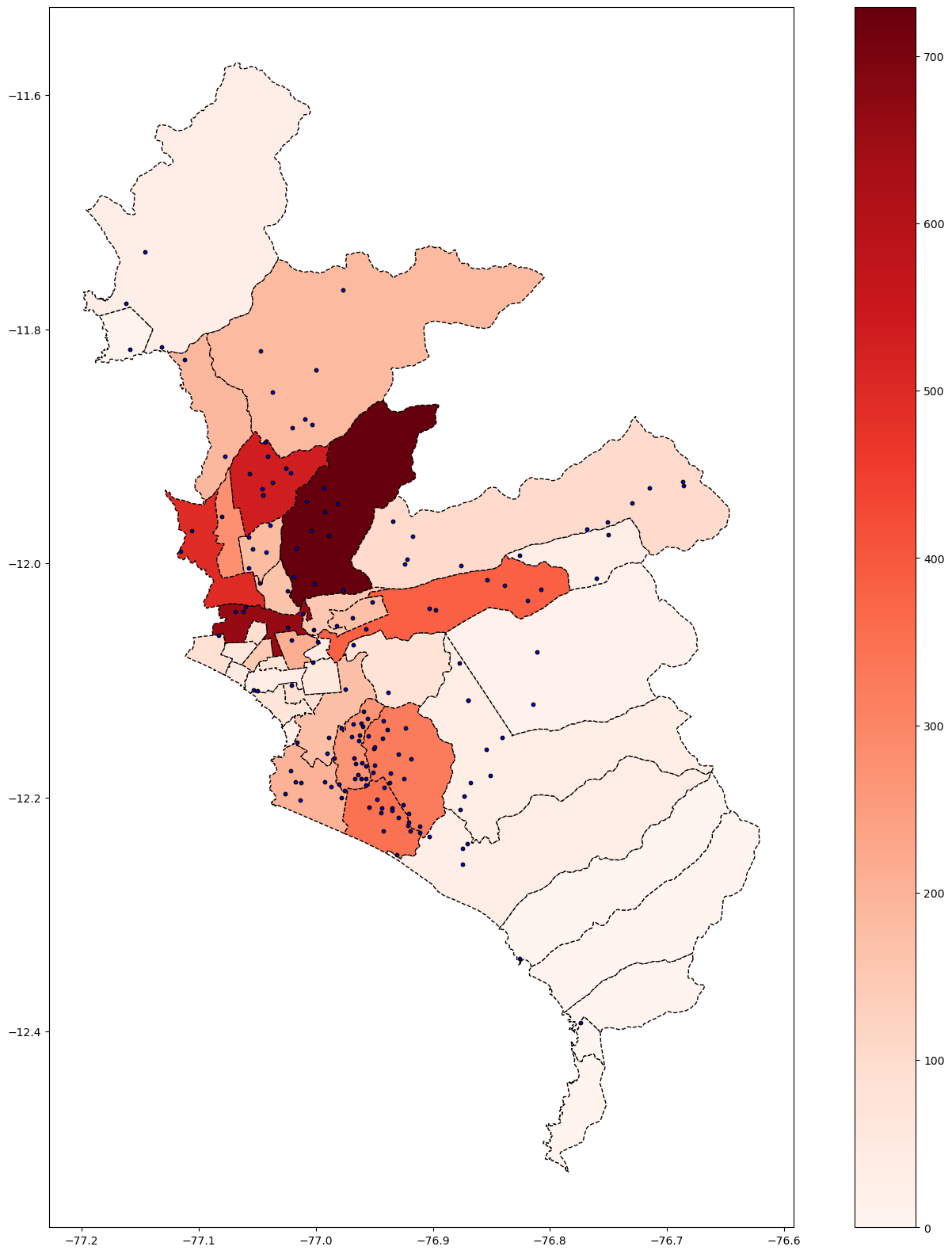
Hospitales Públicos y Postas Médicas#
ax = base_lima.plot(figsize=(25, 25), color='none', edgecolor='black', zorder=3)
PH_lima_postas.plot(color='teal', label = "Postas médicas", markersize=25, ax=ax)
PH_lima_hospitales.plot(color='red', label = "Hospitales públicos", markersize=75, ax=ax)
plt.legend(loc='upper right')
<matplotlib.legend.Legend at 0x1ea28880b20>
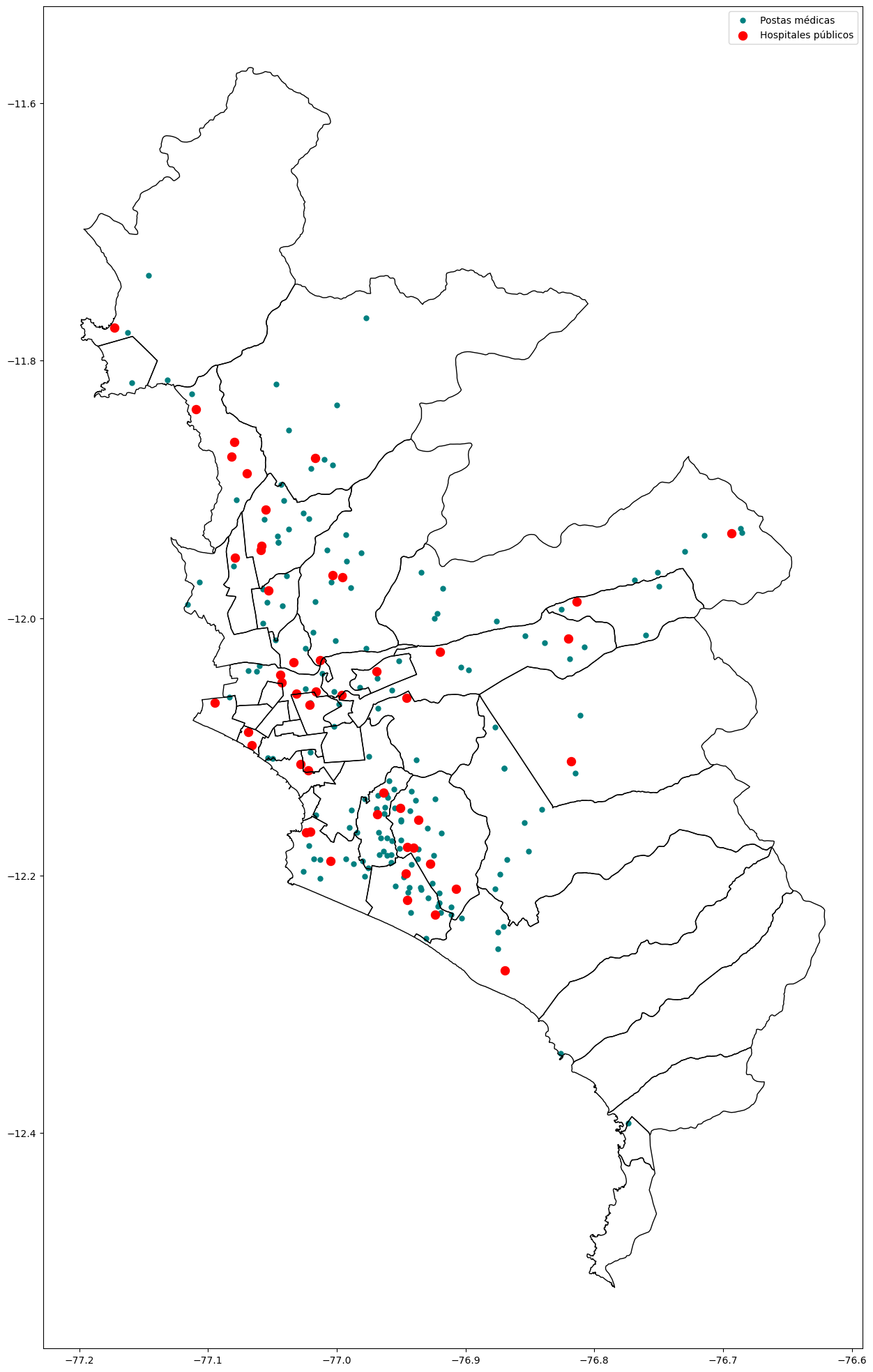
# Covid infection and Postas médicas
fig, ax = plt.subplots(figsize=(20, 20))
#Heatmap Lima and Covid infection
base_lima.plot(column='Casos', cmap='Reds', linestyle='--',
edgecolor='black',
legend = True, ax=ax)
PH_lima_postas.plot(color='black', label = "Postas médicas", markersize=25, ax=ax)
PH_lima_hospitales.plot(color='blue', label = "Hospitales públicos", markersize=75, ax=ax)
plt.legend(loc='upper right')
<matplotlib.legend.Legend at 0x1ea2a4b7e80>
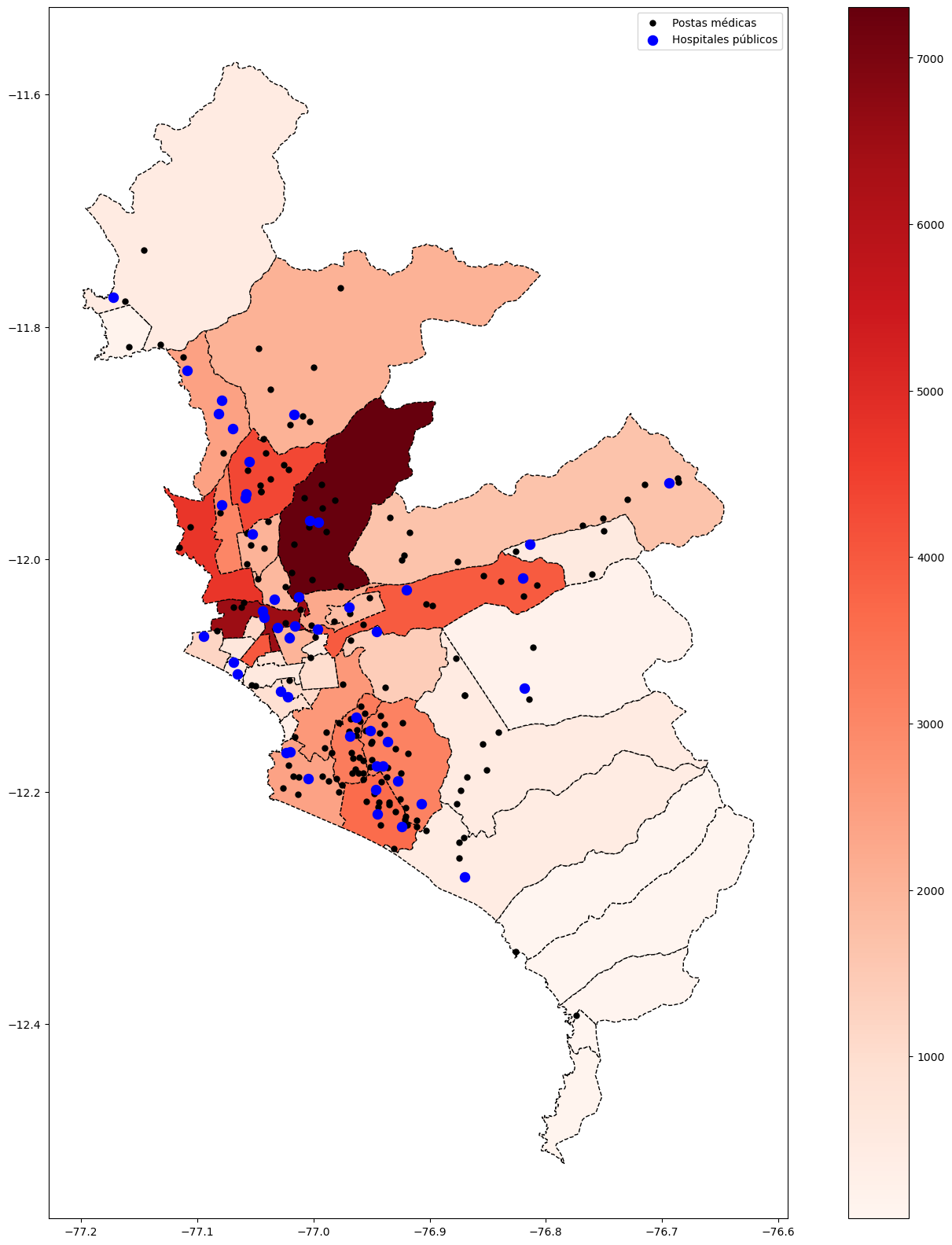
PH_lima_hospitales['CAMAS'].describe
<bound method NDFrame.describe of 2715 NaN
2803 NaN
2832 49.0
2852 NaN
2859 102.0
2862 NaN
2875 NaN
2879 NaN
2887 590.0
2904 NaN
2905 106.0
2912 NaN
2927 NaN
2943 49.0
2955 662.0
2984 NaN
2988 231.0
2997 84.0
3009 NaN
3017 NaN
3023 NaN
3027 NaN
3036 NaN
3040 594.0
3048 NaN
3071 NaN
3077 150.0
3093 6.0
3096 54.0
3161 6.0
3164 NaN
3175 NaN
3204 NaN
3213 NaN
3214 50.0
3216 NaN
3217 NaN
3230 199.0
3238 NaN
3255 NaN
3262 NaN
3269 111.0
3284 NaN
3322 NaN
3363 51.0
3370 NaN
3382 10.0
3508 67.0
7909 57.0
Name: CAMAS, dtype: float64>
# drop raws with nan values
PH_lima_hospitales = PH_lima_hospitales.dropna(subset=['CAMAS'])
#markersize
Relative Size of Markers from panda series#
PH_lima_hospitales["CAMAS"]
2832 49.0
2859 102.0
2887 590.0
2905 106.0
2943 49.0
2955 662.0
2988 231.0
2997 84.0
3040 594.0
3077 150.0
3093 6.0
3096 54.0
3161 6.0
3214 50.0
3230 199.0
3269 111.0
3363 51.0
3382 10.0
3508 67.0
7909 57.0
Name: CAMAS, dtype: float64
# Covid infection and Public Hospital by number of beds
fig, ax = plt.subplots(figsize=(15, 15))
#define the size
markersize=PH_lima_hospitales['CAMAS']
#Heatmap Lima and Covid infection
base_lima.plot(column='Casos', cmap='Reds', linestyle='--',
edgecolor='black',
legend = True, ax=ax)
PH_lima_hospitales.plot( ax=ax, color='blue', label = "Hospitales públicos", markersize = markersize,
edgecolor='black')
plt.legend(loc='upper right')
<matplotlib.legend.Legend at 0x1ea2da469d0>
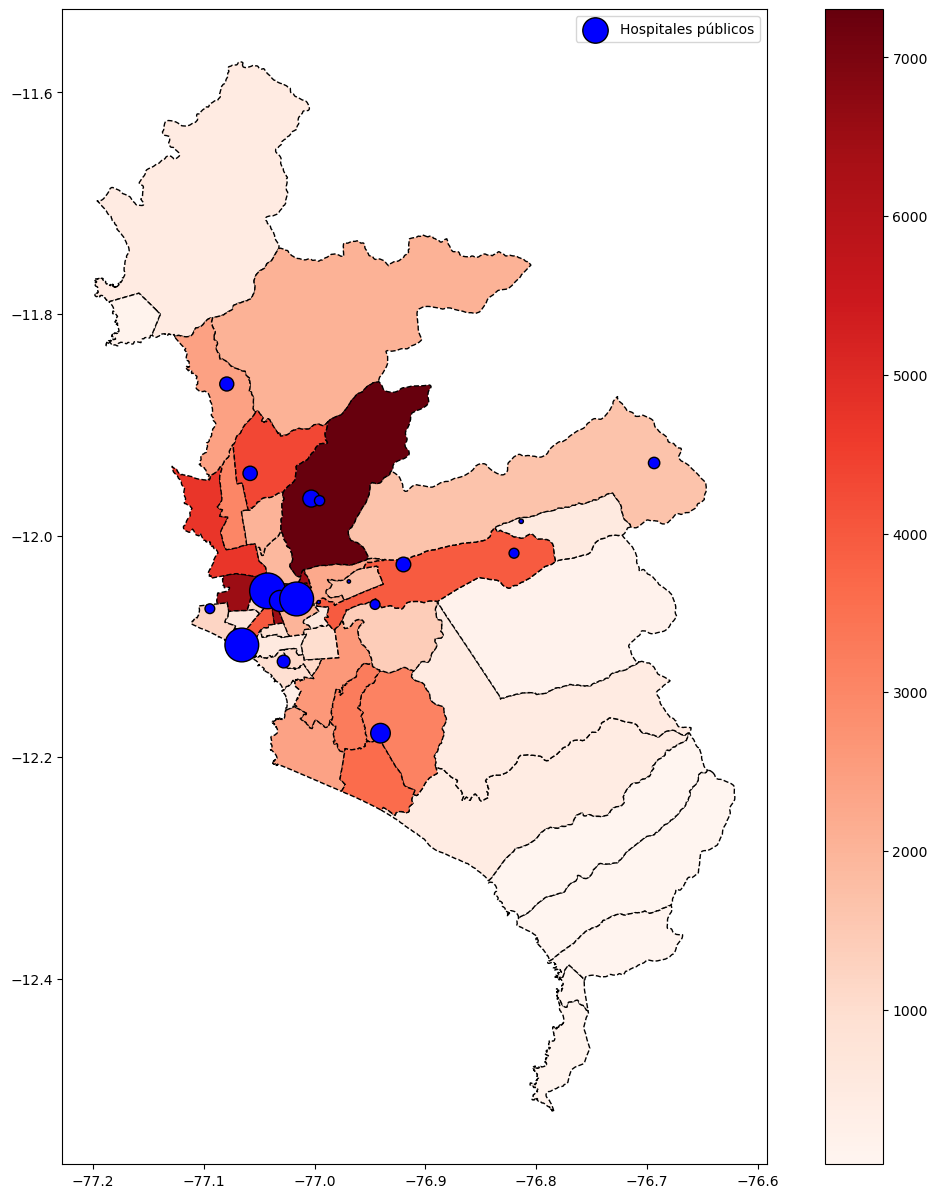
# Covid infection & postas médicas at the national level
gf = base.plot(column='Casos', cmap='Oranges',
figsize=(30, 30),
linestyle='-',
edgecolor='black',
legend = True)
Public_health1.plot(ax = gf, color = 'red', markersize=15)
<AxesSubplot:>
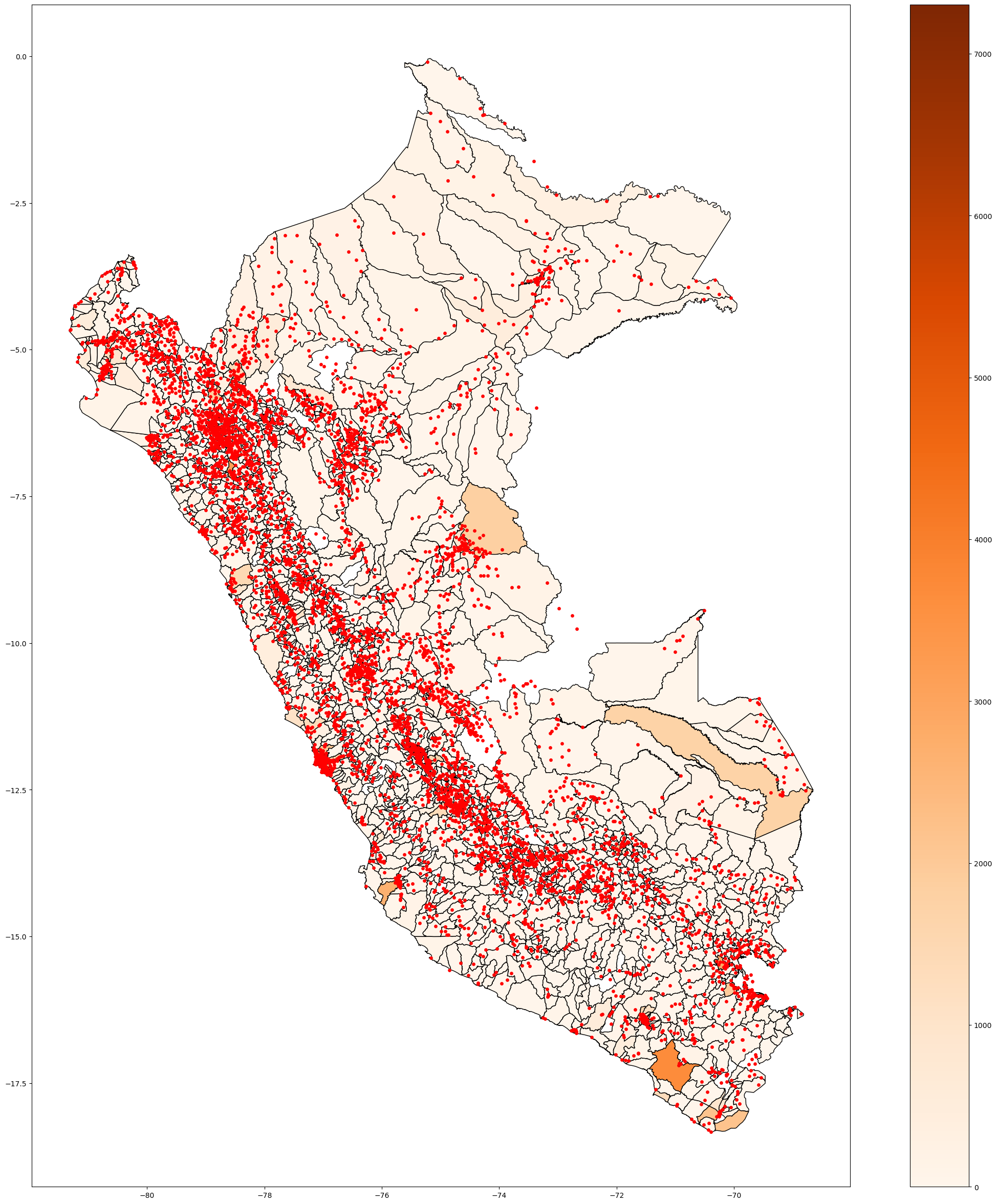
Select Districts#
base_lima = base_lima.set_index("Distrito")
base_lima
| UBIGEO | geometry | Departamento | Provincia | Mes | Año | Casos | Muertes Covid | Muertes totales | centroid | |
|---|---|---|---|---|---|---|---|---|---|---|
| Distrito | ||||||||||
| LURIN | 150119 | POLYGON ((-76.70549 -12.17672, -76.70429 -12.1... | LIMA | LIMA | 8 | 2020 | 452 | 29 | 68 | POINT (-76.80109 -12.23426) |
| ANCON | 150102 | POLYGON ((-77.06517 -11.57512, -77.06505 -11.5... | LIMA | LIMA | 8 | 2020 | 460 | 30 | 51 | POINT (-77.09580 -11.70252) |
| SAN ISIDRO | 150131 | POLYGON ((-77.04859 -12.08504, -77.04762 -12.0... | LIMA | LIMA | 8 | 2020 | 551 | 30 | 86 | POINT (-77.03667 -12.09898) |
| MAGDALENA DEL MAR | 150120 | POLYGON ((-77.07210 -12.08358, -77.06348 -12.0... | LIMA | LIMA | 8 | 2020 | 478 | 49 | 94 | POINT (-77.06669 -12.09384) |
| SAN BORJA | 150130 | POLYGON ((-76.98732 -12.07994, -76.98425 -12.0... | LIMA | LIMA | 8 | 2020 | 1012 | 54 | 123 | POINT (-76.99521 -12.09755) |
| LINCE | 150116 | POLYGON ((-77.02807 -12.07871, -77.02435 -12.0... | LIMA | LIMA | 8 | 2020 | 855 | 76 | 129 | POINT (-77.03567 -12.08579) |
| SANTIAGO DE SURCO | 150140 | POLYGON ((-76.95923 -12.07576, -76.95831 -12.0... | LIMA | LIMA | 8 | 2020 | 2628 | 171 | 394 | POINT (-76.98431 -12.12699) |
| PUEBLO LIBRE | 150121 | POLYGON ((-77.04966 -12.07025, -77.05097 -12.0... | LIMA | LIMA | 8 | 2020 | 633 | 61 | 140 | POINT (-77.06574 -12.07508) |
| SANTA ROSA | 150139 | POLYGON ((-77.14355 -11.79587, -77.14061 -11.7... | LIMA | LIMA | 8 | 2020 | 114 | 9 | 15 | POINT (-77.16325 -11.80501) |
| CARABAYLLO | 150106 | POLYGON ((-76.89995 -11.72979, -76.89921 -11.7... | LIMA | LIMA | 8 | 2020 | 2054 | 183 | 281 | POINT (-76.97291 -11.80738) |
| JESUS MARIA | 150113 | POLYGON ((-77.03811 -12.06768, -77.03629 -12.0... | LIMA | LIMA | 8 | 2020 | 4012 | 147 | 186 | POINT (-77.04821 -12.07811) |
| PUCUSANA | 150124 | POLYGON ((-76.77367 -12.42553, -76.77332 -12.4... | LIMA | LIMA | 8 | 2020 | 90 | 8 | 13 | POINT (-76.77562 -12.46584) |
| SANTA MARIA DEL MAR | 150138 | MULTIPOLYGON (((-76.78117 -12.40917, -76.78144... | LIMA | LIMA | 8 | 2020 | 26 | 0 | 0 | POINT (-76.76740 -12.40858) |
| CHORRILLOS | 150108 | POLYGON ((-77.00745 -12.16033, -77.00781 -12.1... | LIMA | LIMA | 8 | 2020 | 2400 | 203 | 333 | POINT (-77.00589 -12.19265) |
| BARRANCO | 150104 | POLYGON ((-77.01945 -12.13060, -77.01915 -12.1... | LIMA | LIMA | 8 | 2020 | 298 | 26 | 62 | POINT (-77.02086 -12.14403) |
| VILLA MARIA DEL TRIUNFO | 150143 | POLYGON ((-76.91078 -12.12233, -76.91191 -12.1... | LIMA | LIMA | 8 | 2020 | 3123 | 324 | 508 | POINT (-76.91899 -12.17039) |
| SAN JUAN DE MIRAFLORES | 150133 | POLYGON ((-76.95434 -12.11556, -76.95416 -12.1... | LIMA | LIMA | 8 | 2020 | 3251 | 267 | 479 | POINT (-76.96651 -12.15786) |
| MIRAFLORES | 150122 | POLYGON ((-77.02656 -12.10820, -77.02631 -12.1... | LIMA | LIMA | 8 | 2020 | 951 | 66 | 178 | POINT (-77.02893 -12.12091) |
| SURQUILLO | 150141 | POLYGON ((-77.01498 -12.10778, -77.01469 -12.1... | LIMA | LIMA | 8 | 2020 | 1103 | 91 | 170 | POINT (-77.01299 -12.11344) |
| PUNTA NEGRA | 150127 | POLYGON ((-76.64078 -12.21377, -76.64000 -12.2... | LIMA | LIMA | 8 | 2020 | 54 | 3 | 7 | POINT (-76.70943 -12.30363) |
| SAN LUIS | 150134 | POLYGON ((-76.99780 -12.06084, -76.99810 -12.0... | LIMA | LIMA | 8 | 2020 | 567 | 39 | 72 | POINT (-76.99724 -12.07409) |
| SAN MIGUEL | 150136 | POLYGON ((-77.07857 -12.06228, -77.07815 -12.0... | LIMA | LIMA | 8 | 2020 | 1205 | 89 | 190 | POINT (-77.09007 -12.07644) |
| VILLA EL SALVADOR | 150142 | POLYGON ((-76.94232 -12.18247, -76.93731 -12.1... | LIMA | LIMA | 8 | 2020 | 3625 | 346 | 415 | POINT (-76.94528 -12.21928) |
| PUNTA HERMOSA | 150126 | POLYGON ((-76.65267 -12.19256, -76.65207 -12.1... | LIMA | LIMA | 8 | 2020 | 35 | 1 | 7 | POINT (-76.74377 -12.26927) |
| SAN BARTOLO | 150129 | POLYGON ((-76.67773 -12.33800, -76.67787 -12.3... | LIMA | LIMA | 8 | 2020 | 80 | 4 | 10 | POINT (-76.72414 -12.36938) |
| PACHACAMAC | 150123 | POLYGON ((-76.88134 -12.07220, -76.86137 -12.1... | LIMA | LIMA | 8 | 2020 | 464 | 34 | 76 | POINT (-76.80961 -12.15999) |
| LA VICTORIA | 150115 | POLYGON ((-77.01661 -12.05889, -77.01559 -12.0... | LIMA | LIMA | 8 | 2020 | 2003 | 211 | 351 | POINT (-77.01739 -12.07233) |
| LA MOLINA | 150114 | POLYGON ((-76.94006 -12.06405, -76.93942 -12.0... | LIMA | LIMA | 8 | 2020 | 1406 | 81 | 154 | POINT (-76.92573 -12.08809) |
| BRE<d1>A | 150105 | POLYGON ((-77.04206 -12.05348, -77.04198 -12.0... | LIMA | LIMA | 8 | 2020 | 1129 | 95 | 180 | POINT (-77.05059 -12.05916) |
| SANTA ANITA | 150137 | POLYGON ((-76.93981 -12.03913, -76.93796 -12.0... | LIMA | LIMA | 8 | 2020 | 1762 | 169 | 261 | POINT (-76.96314 -12.04321) |
| CIENEGUILLA | 150109 | POLYGON ((-76.72948 -11.99943, -76.72596 -11.9... | LIMA | LIMA | 8 | 2020 | 161 | 12 | 25 | POINT (-76.77752 -12.07646) |
| LIMA | 150101 | POLYGON ((-77.00474 -12.04166, -77.00296 -12.0... | LIMA | LIMA | 8 | 2020 | 6510 | 658 | 666 | POINT (-77.04887 -12.05103) |
| EL AGUSTINO | 150111 | POLYGON ((-76.94408 -12.02075, -76.94348 -12.0... | LIMA | LIMA | 8 | 2020 | 2248 | 153 | 209 | POINT (-76.98740 -12.04010) |
| RIMAC | 150128 | POLYGON ((-77.02034 -12.01783, -77.02032 -12.0... | LIMA | LIMA | 8 | 2020 | 1911 | 163 | 277 | POINT (-77.03268 -12.02358) |
| ATE | 150103 | POLYGON ((-76.83508 -11.99627, -76.83467 -11.9... | LIMA | LIMA | 8 | 2020 | 3969 | 382 | 545 | POINT (-76.87747 -12.03257) |
| CHACLACAYO | 150107 | POLYGON ((-76.72918 -11.96426, -76.72809 -11.9... | LIMA | LIMA | 8 | 2020 | 512 | 31 | 62 | POINT (-76.76695 -11.99164) |
| INDEPENDENCIA | 150112 | POLYGON ((-77.02858 -11.96462, -77.02794 -11.9... | LIMA | LIMA | 8 | 2020 | 2044 | 181 | 277 | POINT (-77.04604 -11.98747) |
| SAN MARTIN DE PORRES | 150135 | POLYGON ((-77.08576 -11.94869, -77.08557 -11.9... | LIMA | LIMA | 8 | 2020 | 4730 | 498 | 768 | POINT (-77.08841 -11.99283) |
| LOS OLIVOS | 150117 | POLYGON ((-77.07044 -11.95013, -77.07008 -11.9... | LIMA | LIMA | 8 | 2020 | 3023 | 279 | 395 | POINT (-77.07437 -11.97242) |
| COMAS | 150110 | POLYGON ((-77.04639 -11.89743, -77.04551 -11.8... | LIMA | LIMA | 8 | 2020 | 4323 | 533 | 753 | POINT (-77.03890 -11.92906) |
| LURIGANCHO | 150118 | POLYGON ((-76.71091 -11.89215, -76.70963 -11.8... | LIMA | LIMA | 8 | 2020 | 1669 | 103 | 172 | POINT (-76.80130 -11.95849) |
| SAN JUAN DE LURIGANCHO | 150132 | POLYGON ((-76.92923 -11.87063, -76.92891 -11.8... | LIMA | LIMA | 8 | 2020 | 7303 | 729 | 1071 | POINT (-76.97149 -11.94596) |
| PUENTE PIEDRA | 150125 | POLYGON ((-77.09076 -11.81966, -77.09066 -11.8... | LIMA | LIMA | 8 | 2020 | 2437 | 189 | 311 | POINT (-77.08665 -11.87212) |
base_lima.buffer(10000)
C:\Users\DELL\AppData\Local\Temp\ipykernel_20856\1713097020.py:1: UserWarning: Geometry is in a geographic CRS. Results from 'buffer' are likely incorrect. Use 'GeoSeries.to_crs()' to re-project geometries to a projected CRS before this operation.
base_lima.buffer(10000)
Distrito
LURIN POLYGON ((-907.44458 9940.34876, -908.51450 99...
ANCON POLYGON ((-10070.83398 214.65757, -10071.54511...
SAN ISIDRO POLYGON ((-6569.98336 7593.29451, -5804.27878 ...
MAGDALENA DEL MAR POLYGON ((-6950.49655 7251.25149, -6191.31465 ...
SAN BORJA POLYGON ((-1770.66790 9843.44877, -791.27992 9...
LINCE POLYGON ((-1448.68595 9893.40233, -503.73208 9...
SANTIAGO DE SURCO POLYGON ((-3326.57958 9440.77630, -3327.93789 ...
PUEBLO LIBRE POLYGON ((6246.29993 7734.88076, 6978.22097 70...
SANTA ROSA POLYGON ((6739.98962 7295.68218, 6744.33819 73...
CARABAYLLO POLYGON ((-824.56307 9955.13861, -824.94665 99...
JESUS MARIA POLYGON ((9774.92742 -1670.75098, 9646.93969 -...
PUCUSANA POLYGON ((-10067.49189 -109.85715, -10076.3141...
SANTA MARIA DEL MAR POLYGON ((-10072.02469 285.02259, -10072.15474...
CHORRILLOS POLYGON ((2625.70000 9605.45191, 2628.36453 96...
BARRANCO POLYGON ((-4122.52578 9133.02542, -3210.01547 ...
VILLA MARIA DEL TRIUNFO POLYGON ((6024.86571 7910.52392, 6622.82282 74...
SAN JUAN DE MIRAFLORES POLYGON ((-673.87963 9963.48472, -674.27124 99...
MIRAFLORES POLYGON ((3063.39167 9476.75000, 3064.95212 94...
SURQUILLO POLYGON ((-1843.75512 9820.79236, -1845.45982 ...
PUNTA NEGRA POLYGON ((4934.72500 8634.06896, 4937.93538 86...
SAN LUIS POLYGON ((253.87111 9967.00974, 254.38285 9982...
SAN MIGUEL POLYGON ((9244.81994 3574.07053, 9256.11314 35...
VILLA EL SALVADOR POLYGON ((-2702.25905 9637.05129, -1723.75830 ...
PUNTA HERMOSA POLYGON ((8411.40078 5274.86226, 8826.69507 45...
SAN BARTOLO POLYGON ((9015.39174 4138.57377, 9020.13227 41...
PACHACAMAC POLYGON ((657.17981 9950.55654, 657.93932 9960...
LA VICTORIA POLYGON ((-1467.67651 9881.87785, -1468.90210 ...
LA MOLINA POLYGON ((-2751.13393 9614.40010, -2753.54201 ...
BRE<d1>A POLYGON ((9837.29920 1294.02360, 9861.22235 10...
SANTA ANITA POLYGON ((9220.46520 3659.63448, 9224.05016 36...
CIENEGUILLA POLYGON ((-1965.55027 9799.12612, -1967.19709 ...
LIMA POLYGON ((9389.02326 3170.20160, 9401.71626 31...
EL AGUSTINO POLYGON ((-445.17756 9971.21950, -445.54509 99...
RIMAC POLYGON ((9165.35017 3805.10518, 9165.72678 38...
ATE POLYGON ((-4911.98416 8728.08993, -4917.60810 ...
CHACLACAYO POLYGON ((7809.82534 6122.11638, 7816.74728 61...
INDEPENDENCIA POLYGON ((8566.30170 4998.70446, 8574.35284 50...
SAN MARTIN DE PORRES POLYGON ((-3092.04918 9521.85319, -3092.29916 ...
LOS OLIVOS POLYGON ((1304.41089 9884.70433, 1305.43231 98...
COMAS POLYGON ((-8873.15610 4734.64793, -8877.50202 ...
LURIGANCHO POLYGON ((-1863.75886 9815.89371, -1865.73751 ...
SAN JUAN DE LURIGANCHO POLYGON ((-4451.28113 8977.85197, -4452.37360 ...
PUENTE PIEDRA POLYGON ((7218.40888 6810.72559, 7226.75949 68...
dtype: geometry
# Get buffered region and centroid points
base_lima["buffered"] = base_lima.buffer(10000)
base_lima['centroid'] = base_lima.centroid
C:\Users\DELL\AppData\Local\Temp\ipykernel_20856\2755669895.py:2: UserWarning: Geometry is in a geographic CRS. Results from 'buffer' are likely incorrect. Use 'GeoSeries.to_crs()' to re-project geometries to a projected CRS before this operation.
base_lima["buffered"] = base_lima.buffer(10000)
C:\Users\DELL\AppData\Local\Temp\ipykernel_20856\2755669895.py:3: UserWarning: Geometry is in a geographic CRS. Results from 'centroid' are likely incorrect. Use 'GeoSeries.to_crs()' to re-project geometries to a projected CRS before this operation.
base_lima['centroid'] = base_lima.centroid
base_lima.buffered.plot()
<AxesSubplot:>
ATE = base_lima[base_lima.UBIGEO == 150103]
ATE.plot(figsize=(10, 10))
<AxesSubplot:>
#Plot selectec districs
ATE = base_lima.loc["ATE", "geometry"]
SJL = base_lima.loc["SAN JUAN DE LURIGANCHO", "geometry"]
SI = base_lima.loc["SAN ISIDRO", "geometry"]
LC = base_lima.loc["LINCE", "geometry"]
gpd.GeoSeries([ATE,SJL,SI,LC]).plot(figsize=(8, 8),linestyle='-',
edgecolor='black')
<AxesSubplot:>
Limits - Boarders#
# Logic operations:
# Lince and San Isidro share limits
LC.touches(SI)
True
LC.touches(ATE)
False
#Distance ATE y (SAN ISIDRO, LINCE)
ATE.distance(SI)
0.021430816237973403
gpd.GeoSeries([ATE, SI]).plot(figsize=(8, 8),linestyle='-',
edgecolor='black')
<AxesSubplot:>
ATE.distance(LC)
0.03353670040995021
# Add line that cross Lince and ATE
ate_point = base_lima['centroid']['ATE']
lc_point = base_lima['centroid']['LINCE']
sb_point = base_lima['centroid']['SAN BORJA']
lr_point = base_lima['centroid']['LURIN']
ac_point = base_lima['centroid']['ANCON']
# Add line that cross Lince and ATE
ate_lince = LineString(zip((ate_point.x , lc_point.x ), (ate_point.y, lc_point.y)))
fig, ax = plt.subplots(figsize=(20,20))
base_lima.plot(color='lightgray', ax=ax, linestyle='-',
edgecolor='black')
gpd.GeoSeries([ate_lince]).plot(color='red', linestyle='--', ax = ax)
<AxesSubplot:>
base_lima['area']=base_lima.area
base_lima
C:\Users\DELL\AppData\Local\Temp\ipykernel_20856\3468981840.py:1: UserWarning: Geometry is in a geographic CRS. Results from 'area' are likely incorrect. Use 'GeoSeries.to_crs()' to re-project geometries to a projected CRS before this operation.
base_lima['area']=base_lima.area
| UBIGEO | geometry | Departamento | Provincia | Mes | Año | Casos | Muertes Covid | Muertes totales | centroid | buffered | area | distance | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Distrito | |||||||||||||
| LURIN | 150119 | POLYGON ((-76.70549 -12.17672, -76.70429 -12.1... | LIMA | LIMA | 8 | 2020 | 452 | 29 | 68 | POINT (-76.80111 -12.23426) | POLYGON ((-907.44458 9940.34876, -908.51450 99... | 0.015586 | 0.334878 |
| ANCON | 150102 | POLYGON ((-77.06517 -11.57512, -77.06505 -11.5... | LIMA | LIMA | 8 | 2020 | 460 | 30 | 51 | POINT (-77.09581 -11.70253) | POLYGON ((-10070.83398 214.65757, -10071.54511... | 0.025751 | 0.273336 |
| SAN ISIDRO | 150131 | POLYGON ((-77.04859 -12.08504, -77.04762 -12.0... | LIMA | LIMA | 8 | 2020 | 551 | 30 | 86 | POINT (-77.03667 -12.09898) | POLYGON ((-6569.98336 7593.29451, -5804.27878 ... | 0.000805 | 0.166317 |
| MAGDALENA DEL MAR | 150120 | POLYGON ((-77.07210 -12.08358, -77.06348 -12.0... | LIMA | LIMA | 8 | 2020 | 478 | 49 | 94 | POINT (-77.06669 -12.09384) | POLYGON ((-6950.49655 7251.25149, -6191.31465 ... | 0.000271 | 0.175867 |
| SAN BORJA | 150130 | POLYGON ((-76.98732 -12.07994, -76.98425 -12.0... | LIMA | LIMA | 8 | 2020 | 1012 | 54 | 123 | POINT (-76.99521 -12.09755) | POLYGON ((-1770.66790 9843.44877, -791.27992 9... | 0.000860 | 0.153424 |
| LINCE | 150116 | POLYGON ((-77.02807 -12.07871, -77.02435 -12.0... | LIMA | LIMA | 8 | 2020 | 855 | 76 | 129 | POINT (-77.03567 -12.08579) | POLYGON ((-1448.68595 9893.40233, -503.73208 9... | 0.000228 | 0.153848 |
| SANTIAGO DE SURCO | 150140 | POLYGON ((-76.95923 -12.07576, -76.95831 -12.0... | LIMA | LIMA | 8 | 2020 | 2628 | 171 | 394 | POINT (-76.98431 -12.12699) | POLYGON ((-3326.57958 9440.77630, -3327.93789 ... | 0.002882 | 0.181479 |
| PUEBLO LIBRE | 150121 | POLYGON ((-77.04966 -12.07025, -77.05097 -12.0... | LIMA | LIMA | 8 | 2020 | 633 | 61 | 140 | POINT (-77.06574 -12.07508) | POLYGON ((6246.29993 7734.88076, 6978.22097 70... | 0.000386 | 0.159857 |
| SANTA ROSA | 150139 | POLYGON ((-77.14355 -11.79587, -77.14061 -11.7... | LIMA | LIMA | 8 | 2020 | 114 | 9 | 15 | POINT (-77.16325 -11.80501) | POLYGON ((6739.98962 7295.68218, 6744.33819 73... | 0.001446 | 0.237993 |
| CARABAYLLO | 150106 | POLYGON ((-76.89995 -11.72979, -76.89921 -11.7... | LIMA | LIMA | 8 | 2020 | 2054 | 183 | 281 | POINT (-76.97293 -11.80738) | POLYGON ((-824.56307 9955.13861, -824.94665 99... | 0.025891 | 0.138590 |
| JESUS MARIA | 150113 | POLYGON ((-77.03811 -12.06768, -77.03629 -12.0... | LIMA | LIMA | 8 | 2020 | 4012 | 147 | 186 | POINT (-77.04821 -12.07811) | POLYGON ((9774.92742 -1670.75098, 9646.93969 -... | 0.000360 | 0.152794 |
| PUCUSANA | 150124 | POLYGON ((-76.77367 -12.42553, -76.77332 -12.4... | LIMA | LIMA | 8 | 2020 | 90 | 8 | 13 | POINT (-76.77563 -12.46584) | POLYGON ((-10067.49189 -109.85715, -10076.3141... | 0.002418 | 0.555547 |
| SANTA MARIA DEL MAR | 150138 | MULTIPOLYGON (((-76.78117 -12.40917, -76.78144... | LIMA | LIMA | 8 | 2020 | 26 | 0 | 0 | POINT (-76.76740 -12.40858) | POLYGON ((-10072.02469 285.02259, -10072.15474... | 0.000644 | 0.505632 |
| CHORRILLOS | 150108 | POLYGON ((-77.00745 -12.16033, -77.00781 -12.1... | LIMA | LIMA | 8 | 2020 | 2400 | 203 | 333 | POINT (-77.00589 -12.19265) | POLYGON ((2625.70000 9605.45191, 2628.36453 96... | 0.003074 | 0.249065 |
| BARRANCO | 150104 | POLYGON ((-77.01945 -12.13060, -77.01915 -12.1... | LIMA | LIMA | 8 | 2020 | 298 | 26 | 62 | POINT (-77.02086 -12.14403) | POLYGON ((-4122.52578 9133.02542, -3210.01547 ... | 0.000228 | 0.204123 |
| VILLA MARIA DEL TRIUNFO | 150143 | POLYGON ((-76.91078 -12.12233, -76.91191 -12.1... | LIMA | LIMA | 8 | 2020 | 3123 | 324 | 508 | POINT (-76.91899 -12.17039) | POLYGON ((6024.86571 7910.52392, 6622.82282 74... | 0.005499 | 0.230486 |
| SAN JUAN DE MIRAFLORES | 150133 | POLYGON ((-76.95434 -12.11556, -76.95416 -12.1... | LIMA | LIMA | 8 | 2020 | 3251 | 267 | 479 | POINT (-76.96651 -12.15786) | POLYGON ((-673.87963 9963.48472, -674.27124 99... | 0.001846 | 0.211952 |
| MIRAFLORES | 150122 | POLYGON ((-77.02656 -12.10820, -77.02631 -12.1... | LIMA | LIMA | 8 | 2020 | 951 | 66 | 178 | POINT (-77.02893 -12.12091) | POLYGON ((3063.39167 9476.75000, 3064.95212 94... | 0.000759 | 0.184133 |
| SURQUILLO | 150141 | POLYGON ((-77.01498 -12.10778, -77.01469 -12.1... | LIMA | LIMA | 8 | 2020 | 1103 | 91 | 170 | POINT (-77.01299 -12.11344) | POLYGON ((-1843.75512 9820.79236, -1845.45982 ... | 0.000384 | 0.172533 |
| PUNTA NEGRA | 150127 | POLYGON ((-76.64078 -12.21377, -76.64000 -12.2... | LIMA | LIMA | 8 | 2020 | 54 | 3 | 7 | POINT (-76.70945 -12.30364) | POLYGON ((4934.72500 8634.06896, 4937.93538 86... | 0.010773 | 0.443395 |
| SAN LUIS | 150134 | POLYGON ((-76.99780 -12.06084, -76.99810 -12.0... | LIMA | LIMA | 8 | 2020 | 567 | 39 | 72 | POINT (-76.99724 -12.07409) | POLYGON ((253.87111 9967.00974, 254.38285 9982... | 0.000289 | 0.130681 |
| SAN MIGUEL | 150136 | POLYGON ((-77.07857 -12.06228, -77.07815 -12.0... | LIMA | LIMA | 8 | 2020 | 1205 | 89 | 190 | POINT (-77.09007 -12.07644) | POLYGON ((9244.81994 3574.07053, 9256.11314 35... | 0.000796 | 0.176307 |
| VILLA EL SALVADOR | 150142 | POLYGON ((-76.94232 -12.18247, -76.93731 -12.1... | LIMA | LIMA | 8 | 2020 | 3625 | 346 | 415 | POINT (-76.94527 -12.21928) | POLYGON ((-2702.25905 9637.05129, -1723.75830 ... | 0.002811 | 0.274567 |
| PUNTA HERMOSA | 150126 | POLYGON ((-76.65267 -12.19256, -76.65207 -12.1... | LIMA | LIMA | 8 | 2020 | 35 | 1 | 7 | POINT (-76.74379 -12.26927) | POLYGON ((8411.40078 5274.86226, 8826.69507 45... | 0.010132 | 0.395445 |
| SAN BARTOLO | 150129 | POLYGON ((-76.67773 -12.33800, -76.67787 -12.3... | LIMA | LIMA | 8 | 2020 | 80 | 4 | 10 | POINT (-76.72414 -12.36938) | POLYGON ((9015.39174 4138.57377, 9020.13227 41... | 0.004458 | 0.490367 |
| PACHACAMAC | 150123 | POLYGON ((-76.88134 -12.07220, -76.86137 -12.1... | LIMA | LIMA | 8 | 2020 | 464 | 34 | 76 | POINT (-76.80962 -12.15999) | POLYGON ((657.17981 9950.55654, 657.93932 9960... | 0.015438 | 0.268347 |
| LA VICTORIA | 150115 | POLYGON ((-77.01661 -12.05889, -77.01559 -12.0... | LIMA | LIMA | 8 | 2020 | 2003 | 211 | 351 | POINT (-77.01739 -12.07233) | POLYGON ((-1467.67651 9881.87785, -1468.90210 ... | 0.000759 | 0.134441 |
| LA MOLINA | 150114 | POLYGON ((-76.94006 -12.06405, -76.93942 -12.0... | LIMA | LIMA | 8 | 2020 | 1406 | 81 | 154 | POINT (-76.92573 -12.08809) | POLYGON ((-2751.13393 9614.40010, -2753.54201 ... | 0.004046 | 0.149307 |
| BRE<d1>A | 150105 | POLYGON ((-77.04206 -12.05348, -77.04198 -12.0... | LIMA | LIMA | 8 | 2020 | 1129 | 95 | 180 | POINT (-77.05059 -12.05916) | POLYGON ((9837.29920 1294.02360, 9861.22235 10... | 0.000267 | 0.138095 |
| SANTA ANITA | 150137 | POLYGON ((-76.93981 -12.03913, -76.93796 -12.0... | LIMA | LIMA | 8 | 2020 | 1762 | 169 | 261 | POINT (-76.96314 -12.04321) | POLYGON ((9220.46520 3659.63448, 9224.05016 36... | 0.000886 | 0.097599 |
| CIENEGUILLA | 150109 | POLYGON ((-76.72948 -11.99943, -76.72596 -11.9... | LIMA | LIMA | 8 | 2020 | 161 | 12 | 25 | POINT (-76.77753 -12.07646) | POLYGON ((-1965.55027 9799.12612, -1967.19709 ... | 0.018677 | 0.233781 |
| LIMA | 150101 | POLYGON ((-77.00474 -12.04166, -77.00296 -12.0... | LIMA | LIMA | 8 | 2020 | 6510 | 658 | 666 | POINT (-77.04887 -12.05103) | POLYGON ((9389.02326 3170.20160, 9401.71626 31... | 0.001786 | 0.130485 |
| EL AGUSTINO | 150111 | POLYGON ((-76.94408 -12.02075, -76.94348 -12.0... | LIMA | LIMA | 8 | 2020 | 2248 | 153 | 209 | POINT (-76.98740 -12.04010) | POLYGON ((-445.17756 9971.21950, -445.54509 99... | 0.001111 | 0.095466 |
| RIMAC | 150128 | POLYGON ((-77.02034 -12.01783, -77.02032 -12.0... | LIMA | LIMA | 8 | 2020 | 1911 | 163 | 277 | POINT (-77.03268 -12.02358) | POLYGON ((9165.35017 3805.10518, 9165.72678 38... | 0.001001 | 0.098832 |
| ATE | 150103 | POLYGON ((-76.83508 -11.99627, -76.83467 -11.9... | LIMA | LIMA | 8 | 2020 | 3969 | 382 | 545 | POINT (-76.87748 -12.03257) | POLYGON ((-4911.98416 8728.08993, -4917.60810 ... | 0.007058 | 0.127825 |
| CHACLACAYO | 150107 | POLYGON ((-76.72918 -11.96426, -76.72809 -11.9... | LIMA | LIMA | 8 | 2020 | 512 | 31 | 62 | POINT (-76.76695 -11.99164) | POLYGON ((7809.82534 6122.11638, 7816.74728 61... | 0.003553 | 0.209580 |
| INDEPENDENCIA | 150112 | POLYGON ((-77.02858 -11.96462, -77.02794 -11.9... | LIMA | LIMA | 8 | 2020 | 2044 | 181 | 277 | POINT (-77.04604 -11.98747) | POLYGON ((8566.30170 4998.70446, 8574.35284 50... | 0.001337 | 0.085326 |
| SAN MARTIN DE PORRES | 150135 | POLYGON ((-77.08576 -11.94869, -77.08557 -11.9... | LIMA | LIMA | 8 | 2020 | 4730 | 498 | 768 | POINT (-77.08841 -11.99283) | POLYGON ((-3092.04918 9521.85319, -3092.29916 ... | 0.003015 | 0.125958 |
| LOS OLIVOS | 150117 | POLYGON ((-77.07044 -11.95013, -77.07008 -11.9... | LIMA | LIMA | 8 | 2020 | 3023 | 279 | 395 | POINT (-77.07437 -11.97242) | POLYGON ((1304.41089 9884.70433, 1305.43231 98... | 0.001512 | 0.106220 |
| COMAS | 150110 | POLYGON ((-77.04639 -11.89743, -77.04551 -11.8... | LIMA | LIMA | 8 | 2020 | 4323 | 533 | 753 | POINT (-77.03890 -11.92906) | POLYGON ((-8873.15610 4734.64793, -8877.50202 ... | 0.004055 | 0.069492 |
| LURIGANCHO | 150118 | POLYGON ((-76.71091 -11.89215, -76.70963 -11.8... | LIMA | LIMA | 8 | 2020 | 1669 | 103 | 172 | POINT (-76.80133 -11.95848) | POLYGON ((-1863.75886 9815.89371, -1865.73751 ... | 0.020338 | 0.170624 |
| SAN JUAN DE LURIGANCHO | 150132 | POLYGON ((-76.92923 -11.87063, -76.92891 -11.8... | LIMA | LIMA | 8 | 2020 | 7303 | 729 | 1071 | POINT (-76.97149 -11.94597) | POLYGON ((-4451.28113 8977.85197, -4452.37360 ... | 0.011652 | 0.000000 |
| PUENTE PIEDRA | 150125 | POLYGON ((-77.09076 -11.81966, -77.09066 -11.8... | LIMA | LIMA | 8 | 2020 | 2437 | 189 | 311 | POINT (-77.08665 -11.87213) | POLYGON ((7218.40888 6810.72559, 7226.75949 68... | 0.004193 | 0.136796 |
#Distamce between San Juan de Lurigancho to other districs
SJL_point = base_lima['centroid']["SAN JUAN DE LURIGANCHO"]
base_lima['distance'] = base_lima['centroid'].distance(SJL_point)
C:\Users\DELL\AppData\Local\Temp\ipykernel_20856\3552139678.py:5: UserWarning: Geometry is in a geographic CRS. Results from 'distance' are likely incorrect. Use 'GeoSeries.to_crs()' to re-project geometries to a projected CRS before this operation.
base_lima['distance'] = base_lima['centroid'].distance(SJL_point)
base_lima['distance'].sort_values(ascending=False)
Distrito
PUCUSANA 0.555547
SANTA MARIA DEL MAR 0.505632
SAN BARTOLO 0.490367
PUNTA NEGRA 0.443395
PUNTA HERMOSA 0.395445
LURIN 0.334878
VILLA EL SALVADOR 0.274567
ANCON 0.273336
PACHACAMAC 0.268347
CHORRILLOS 0.249065
SANTA ROSA 0.237993
CIENEGUILLA 0.233781
VILLA MARIA DEL TRIUNFO 0.230486
SAN JUAN DE MIRAFLORES 0.211952
CHACLACAYO 0.209580
BARRANCO 0.204123
MIRAFLORES 0.184133
SANTIAGO DE SURCO 0.181479
SAN MIGUEL 0.176307
MAGDALENA DEL MAR 0.175867
SURQUILLO 0.172533
LURIGANCHO 0.170624
SAN ISIDRO 0.166317
PUEBLO LIBRE 0.159857
LINCE 0.153848
SAN BORJA 0.153424
JESUS MARIA 0.152794
LA MOLINA 0.149307
CARABAYLLO 0.138590
BRE<d1>A 0.138095
PUENTE PIEDRA 0.136796
LA VICTORIA 0.134441
SAN LUIS 0.130681
LIMA 0.130485
ATE 0.127825
SAN MARTIN DE PORRES 0.125958
LOS OLIVOS 0.106220
RIMAC 0.098832
SANTA ANITA 0.097599
EL AGUSTINO 0.095466
INDEPENDENCIA 0.085326
COMAS 0.069492
SAN JUAN DE LURIGANCHO 0.000000
Name: distance, dtype: float64
base_lima['distance'].sort_values(ascending=False).plot.bar(figsize=(12, 7), color = 'red', edgecolor='black')
<AxesSubplot:xlabel='Distrito'>
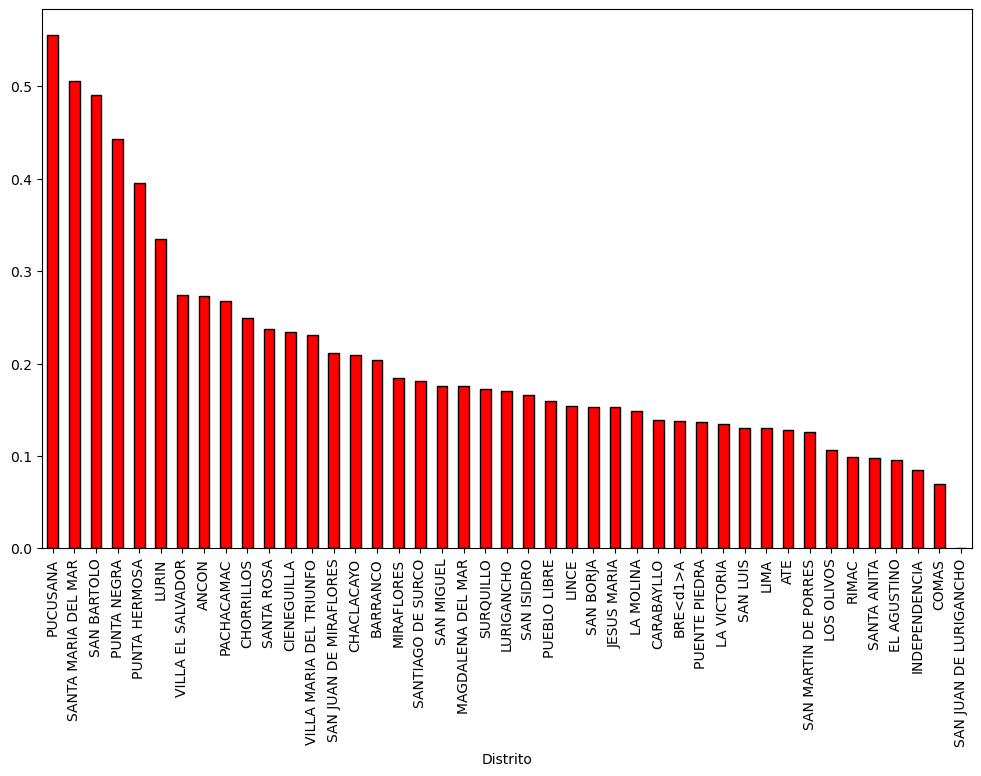
ate_point.type
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
~\AppData\Local\Temp\ipykernel_20856\1350904785.py in <module>
----> 1 ate_point.type
NameError: name 'ate_point' is not defined
# SJL and other districs
d1 = LineString(zip((SJL_point.x , lc_point.x ), (SJL_point.y, lc_point.y)))
d2 = LineString(zip((SJL_point.x , ate_point.x ), (SJL_point.y, ate_point.y)))
d3 = LineString(zip((SJL_point.x , sb_point.x ), (SJL_point.y, sb_point.y)))
d4 = LineString(zip((SJL_point.x , lr_point.x ), (SJL_point.y, lr_point.y)))
d5 = LineString(zip((SJL_point.x , ac_point.x ), (SJL_point.y, ac_point.y)))
fig, ax = plt.subplots(figsize=(20,20))
base_lima.plot(color='white', ax=ax, linestyle='-',
edgecolor='black')
for i in range(1,6):
gpd.GeoSeries(globals()[f'd{i}']).plot(color='red', linestyle='--', ax = ax, linewidth = 2.5)
ArcGis Operations in Geopandas#
dist_shp = gpd.read_file(r'../_data/shape_file/DISTRITOS.shp')
Get Centroids#
Get the centroids of each geometry (district).
dist_shp.crs
<Geographic 2D CRS: EPSG:4326>
Name: WGS 84
Axis Info [ellipsoidal]:
- Lat[north]: Geodetic latitude (degree)
- Lon[east]: Geodetic longitude (degree)
Area of Use:
- name: World
- bounds: (-180.0, -90.0, 180.0, 90.0)
Datum: World Geodetic System 1984
- Ellipsoid: WGS 84
- Prime Meridian: Greenwich
dist_shp.to_crs(24891).centroid.to_crs( 4326 )
0 POINT (-75.46643 -9.63426)
1 POINT (-74.86630 -9.00133)
2 POINT (-75.04801 -8.82493)
3 POINT (-75.27987 -8.95039)
4 POINT (-75.04902 -8.64129)
...
1868 POINT (-76.03798 -9.20808)
1869 POINT (-76.00134 -9.08221)
1870 POINT (-75.98295 -8.98489)
1871 POINT (-70.12642 -15.40623)
1872 POINT (-77.44521 -11.00357)
Length: 1873, dtype: geometry
dist_shp.crs
<Geographic 2D CRS: EPSG:4326>
Name: WGS 84
Axis Info [ellipsoidal]:
- Lat[north]: Geodetic latitude (degree)
- Lon[east]: Geodetic longitude (degree)
Area of Use:
- name: World
- bounds: (-180.0, -90.0, 180.0, 90.0)
Datum: World Geodetic System 1984
- Ellipsoid: WGS 84
- Prime Meridian: Greenwich
We get warnings since we need to reproject this shapefile object to a CRS in meters. This CRS should be a plane area to the specific zone of the shapefile (more info). In the case of Peru, we use 24891 (more info).
projected_dist_shp = dist_shp.to_crs( epsg = 24891 )
projected_dist_shp[ 'centroids'] = projected_dist_shp.centroid
Then, we reproject again the centroids to the original CRS.
dist_shp[ 'Centroid_Latitude' ] = projected_dist_shp[ 'centroids' ].to_crs( epsg = 4326 ).y
dist_shp[ 'Centroid_Longitude' ] = projected_dist_shp[ 'centroids' ].to_crs( epsg = 4326 ).x
Intersection#
health_centers = pd.read_csv( r"../_data\salud_places_peru.csv")
ht1 = health_centers[ health_centers.latitud != 0 ]
# Make ht1 a geopandas
ht1_geo = gpd.GeoDataFrame( ht1 ,
geometry= gpd.points_from_xy(
ht1.longitud,
ht1.latitud))
Check CRS
We found that geolocation is recognized by google maps. We set the CRS that correspond to google maps.
ht1_geo_crs = ht1_geo.set_crs( epsg = 4326 )
Find Ubigeo for each point
# Just take 10% of observations
ht2_geo = ht1_geo_crs.sample( frac = 0.10 )
# Intersect with maps
intersected_ht = gpd.overlay( ht2_geo , maps , how = 'intersection' )
ht2_geo.columns
Index(['id_eess', 'codigo_renaes', 'categoria', 'nombre', 'diresa', 'red',
'direccion', 'longitud', 'latitud', 'id_ubigeo', 'geometry'],
dtype='object')
intersected_ht
| id_eess | codigo_renaes | categoria | nombre | diresa | red | direccion | longitud | latitud | id_ubigeo | UBIGEO | geometry | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 10033 | 19335 | I-3 | POLICLINICO BANDA SHILCAYO | SAN MARTIN | NO PERTENECE A NINGUNA RED | JIRÓN JIRON TARAPOTO 382 BANDA DE SHILCAYO DIS... | -76.346362 | -6.492716 | 1807 | 220909 | POINT (-76.34636 -6.49272) |
| 1 | 11895 | 6367 | I-1 | LA UNION (SANIRARCA) | SAN MARTIN | SAN MARTIN | AVENIDA LA UNION ACSERIO LA UNION AV. LA UNION... | -76.304616 | -6.504808 | 1807 | 220909 | POINT (-76.30462 -6.50481) |
| 2 | 12117 | 18667 | II-E | CLINICA ARROYO | LIMA DIRIS CENTRO | NO PERTENECE A NINGUNA RED | AVENIDA AV. PABLO CARRIQUIRY 798 CORPAC SAN IS... | -77.018614 | -12.099788 | 1311 | 150131 | POINT (-77.01861 -12.09979) |
| 3 | 20464 | 17172 | I-3 | IMM S.A.C. | LIMA DIRIS CENTRO | NO PERTENECE A NINGUNA RED | AVENIDA AV. JAVIER PRADO ESTE 1476 DISTRITO SA... | -77.011404 | -12.089537 | 1311 | 150131 | POINT (-77.01140 -12.08954) |
| 4 | 23555 | 28906 | I-1 | CONSULTORIO ODONTOLOGICO FRANCESQUI | LIMA DIRIS CENTRO | NO PERTENECE A NINGUNA RED | AVENIDA PAZ SOLDAN 170 INT 601 606 DISTRITO SA... | -77.033371 | -12.097050 | 1311 | 150131 | POINT (-77.03337 -12.09705) |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 1539 | 9567 | 488 | I-1 | SAN ANDRES | JUNIN | SATIPO | OTROS COMUNIDAD SAN ANDRES DISTRITO COVIRIALI ... | -74.688297 | -11.329232 | 1124 | 120602 | POINT (-74.68830 -11.32923) |
| 1540 | 2222 | 28935 | I-1 | SHIPETIARI | MADRE DE DIOS | MADRE DE DIOS | CAMINO RURAL Comunidad Shipetiari DISTRITO MAN... | -70.908723 | -12.252546 | 1510 | 170202 | POINT (-70.90872 -12.25255) |
| 1541 | 27640 | 28541 | SD | LABORATORIO DE ANÁLISIS CLÍNICO "VIRGEN DE LOU... | LAMBAYEQUE | NO PERTENECE A NINGUNA RED | JIRÓN CONQUISTA NÚMERO 360 DISTRITO JOSE LEONA... | -79.836558 | -6.760543 | 1247 | 140105 | POINT (-79.83656 -6.76054) |
| 1542 | 16383 | 5296 | II-1 | HOSPITAL DISTRITAL DE PACASMAYO | LA LIBERTAD | RED PACASMAYO | JIRÓN JIRON RAZURI S/N S/N JIRON RAZURI S/N PA... | -79.569327 | -7.398782 | 1205 | 130704 | POINT (-79.56933 -7.39878) |
| 1543 | 4664 | 2681 | I-1 | SAN ANTONIO | APURIMAC | ABANCAY | OTROS PANAMERICANA ABANCAY - CUSCO S/N S/N PAN... | -72.856895 | -13.605669 | 259 | 30109 | POINT (-72.85690 -13.60567) |
1544 rows × 12 columns
Aggregation#
We have shapefiles at district levels. It is possible to aggregate districts at Department or Province level.
dist_shp.plot()
<AxesSubplot:>
prov_shp = dist_shp.dissolve( by = 'IDPROV' )
fig, ax = plt.subplots( figsize = ( 12, 15 ) )
dist_shp.plot( ax = ax )
<AxesSubplot:>
fig, ax = plt.subplots( figsize = ( 12, 15 ) )
prov_shp.plot( ax = ax )
<AxesSubplot:>
dpt_shp = dist_shp.dissolve( by = 'IDDPTO' )
fig, ax = plt.subplots( figsize = ( 12, 15 ) )
dpt_shp.plot( ax = ax )
<AxesSubplot:>
fig, ax = plt.subplots( figsize = ( 12, 15 ) )
dist_shp.plot( ax = ax )
dpt_shp.plot( ax = ax, facecolor="none", edgecolor='black', lw=0.7 )
<AxesSubplot:>
Checking CRS#
# Border of Switzerland
ch_borders = gpd.read_file( r'../_data/eu_maps/CHE_adm0.shp')
# Border of Germany
de_borders = gpd.read_file( r'../_data/eu_maps/gadm36_DEU_0.shp')
# Switzerland lakes shape files
ch_lakes = gpd.GeoDataFrame.from_file( r'../_data/eu_maps/g2s15.shp')
# muni
Geo_muni = gpd.read_file( r"../_data/eu_maps/VG250_GEM.shp" )
Plots with different CRS
fig = plt.figure(figsize=(40, 20), dpi=100)
ax = fig.add_subplot(111, frame_on = False )
# German Plot
Geo_muni.plot( ax = ax )
# switzerland borders
ch_borders.geometry.boundary.plot(color = 'black', ax=ax, edgecolor='0.2', linewidth=1.5)
# # Swiss lakes
ch_lakes.plot(color='lightblue' , ax=ax, edgecolor='0.2', linewidth=0.4, legend=True)
ax.axis('off')
# # define coordinates for the plot area
# ax.set_xlim(455869.5728, 927076.5648)
# #ax.set_ylim( auto = True)
# ax.set_ylim(60443.1884, 529941.7864)
# Save in pdf
# plt.savefig('maps/inventors_BW_bar_legend.pdf')
(-46058.36469219931, 967356.691928138, -305024.0777151486, 6406513.607306512)

Check CRS
shp_files = [ ch_borders, de_borders, ch_lakes, Geo_muni ]
for shp in shp_files:
print( shp.crs )
#cada informacion esta por su CRS, debe cambiarse
epsg:4326
epsg:4326
epsg:21781
epsg:25832
Reproject CRS
Geo_muni_crs = Geo_muni.to_crs( 21781 )
ch_borders_crs = ch_borders.to_crs( 21781 )
fig = plt.figure(figsize=(40, 20), dpi=100)
ax = fig.add_subplot(111, frame_on = False )
# German Plot
Geo_muni_crs.plot( ax = ax )
# switzerland borders
ch_borders_crs.geometry.boundary.plot(color = 'black', ax=ax, edgecolor='0.2', linewidth=1.5)
# # Swiss lakes
ch_lakes.plot(color='lightblue' , ax=ax, edgecolor='0.2', linewidth=0.4, legend=True)
ax.axis('off')
# # define coordinates for the plot area
# ax.set_xlim(455869.5728, 927076.5648)
#ax.set_ylim( auto = True)
# ax.set_ylim(60443.1884, 529941.7864)
# Save in pdf
# plt.savefig('maps/inventors_BW_bar_legend.pdf')
(453126.0049465788, 1163640.4776774019, 11798.918320549732, 1157415.473468453)
References:#
Geopandas#
https://geopandas.org/en/stable/docs/user_guide/set_operations.html
https://geopandas.org/en/stable/docs/user_guide/aggregation_with_dissolve.html
https://geopandas.org/en/stable/docs/user_guide/data_structures.html
Coordinate Reference System#
https://geopandas.org/en/stable/docs/user_guide/projections.html
Types of projections#
Universal Transverse Mercator (UTM)#
https://manifold.net/doc/mfd9/universal_transverse_mercator_projection.htm

