12. Causal Inference#
import pandas as pd
from pandas import Series, DataFrame
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
import seaborn as sns
import chardet
import json
import graphviz as gr
import pickle
import econml ## install package using cmd
import dowhy ## install using cmd
from dowhy import CausalModel
from IPython.display import Image, display
import statistics
from scipy.stats import shapiro
import scipy.stats as stats
from scipy.stats import mannwhitneyu, ks_2samp
import warnings
from sklearn.exceptions import ConvergenceWarning
warnings.simplefilter(action='ignore', category=FutureWarning)
warnings.simplefilter("ignore", category=ConvergenceWarning)
from termcolor import colored
from sklearn import datasets
import statsmodels.formula.api as sm
import econtools.metrics as mt
Causal inference is the leveraging of theory and deep knowledge of institutional details to estimate the impact of events and choices on a given outcome of interest. In the field of economics, we find data from experimental studies or observational data. Each of them presents its own challenges to estimate unbiased causal effect.
1. The challenging to identify causal effect: correlation is not causation#
2. Potencial outcomes \(Y_i^1 \ \ Y_i^0 \)#
\(D_i\) is a binary variable that takes on a value of 1 if a particular unit i receives the treatment and a 0 if it does not. Each unit will have two potential outcomes, but only one observed outcome. Potential outcomes are defined as \(Y_i^1\) if unit received the treatment and as \(Y_i^0\) if the unit did not.
A unit’s observable outcome is a function of its potential outcomes determined according to the switching equation
The unit-specific treatment effect as the difference between the two states of the world:
This requires knowing two states of the world, but we observe only one. Hence, we cannot calculate the unit treatment effect: “counterfactual problem”.
2.1 Average treatment effect#
Difference on population expected means in two states of the world.
We have to estimate ATE because we just one state if the world. We need to find a valid counterfactual.
How estiamte ATE#
2.1.1 Independece Asumption#
Potencial outcome are not correlated to treatment. This eliminates selection bias. In other word, treated and untreated unites are comparable.
2.1.2 SUTVA asumption ( stable unit treatment value assumption)#
It implies two things: the treatment is homogeneous between the treated units and there is no spillobes effect. That is, there are no untreated units that receive treatment
2.2 Random Control Trails (RCT)#
Randomised Experiments. Randomised experiments consist of randomly assigning individuals in a population to a treatment or to a control group.
Randomisation annihilates bias by making the potential outcomes independent of the treatment. Both groups are comparable into observable and non-observable variables.
2.3 Internal and external validity:#
Random selection keeps características from population (external validity). In a similar way, RCT keeps treatment and control group caracteristics (internal validity). Once treatment, both groups are affected by same external factors. Treatment is the unique difference in both groups. RCT overcome selection bias: accurate estimate of contrafactual.
External validity let to extrapolate or generalized conclusion from sample evaluation to la población de unidades elegibles or targeret popultaion.
3 A/B test#
Randomized experimentation process where in two or more versions of a variable (web page, page element, etc.) are shown to different segments of website visitors at the same time to determine which version leaves the maximum impact and drive business metrics.
A/B test application#
An ecommerce company used the following strategy: assign discount benefits on online purchases to its customers randomly. The notice of benefits will appear in the user’s virtual account. This strategy is applied for one year.
The objective of the work is to evaluate the causal effect of the marketing strategy. Does the strategy manage to increase sales? Does the marketing strategy increase sales in a sustained manner?
# Uplaoad base from marketing strategies
base = open(r'../_data/ABtest/abtest.csv', 'rb').read()
det = chardet.detect(base)
charenc = det['encoding']
base1 = pd.read_csv( r'../_data/ABtest/abtest.csv', encoding = charenc)
base1
| user_id | signup_month | month | spend | treatment | |
|---|---|---|---|---|---|
| 0 | 0 | 8 | 1 | 511 | True |
| 1 | 0 | 8 | 2 | 471 | True |
| 2 | 0 | 8 | 3 | 465 | True |
| 3 | 0 | 8 | 4 | 479 | True |
| 4 | 0 | 8 | 5 | 429 | True |
| ... | ... | ... | ... | ... | ... |
| 119995 | 9999 | 8 | 8 | 399 | True |
| 119996 | 9999 | 8 | 9 | 488 | True |
| 119997 | 9999 | 8 | 10 | 524 | True |
| 119998 | 9999 | 8 | 11 | 494 | True |
| 119999 | 9999 | 8 | 12 | 509 | True |
120000 rows × 5 columns
| user_id | signup_month | month | spend | treatment | |
|---|---|---|---|---|---|
| 0 | 0 | 8 | 1 | 511 | True |
| 1 | 0 | 8 | 2 | 471 | True |
| 2 | 0 | 8 | 3 | 465 | True |
| 3 | 0 | 8 | 4 | 479 | True |
| 4 | 0 | 8 | 5 | 429 | True |
| ... | ... | ... | ... | ... | ... |
| 119995 | 9999 | 8 | 8 | 399 | True |
| 119996 | 9999 | 8 | 9 | 488 | True |
| 119997 | 9999 | 8 | 10 | 524 | True |
| 119998 | 9999 | 8 | 11 | 494 | True |
| 119999 | 9999 | 8 | 12 | 509 | True |
120000 rows × 5 columns
base = base1.replace({True: 1, False: 0})
base.dtypes
user_id int64
signup_month int64
month int64
spend int64
treatment int64
dtype: object
user_id int64
signup_month int64
month int64
spend int64
treatment int64
dtype: object
# The function and bucle allows to identify the benefits per month and accumulated sales.
def causal_signup_month(a,i,j):
data = a[(a.signup_month == i) | (a.signup_month == 0)]
data = data[(data.month >= i) & (data.month <= i+j)]
data = data.groupby(['user_id'], as_index=False)['spend','treatment'].sum()
data['treatment'] = data['treatment']/(j+1)
return data
for j in range(1,12):
for i in range(1,12):
globals()[f'base_{i}_{j+1}'] = causal_signup_month(base,i,j)
# total sales under two months of treatment for the beneficiary clients on (February)
# sales under treatment (February and march)
base_2_2
| user_id | spend | treatment | |
|---|---|---|---|
| 0 | 1 | 926 | 0.0 |
| 1 | 2 | 922 | 0.0 |
| 2 | 3 | 1010 | 1.0 |
| 3 | 4 | 958 | 0.0 |
| 4 | 9 | 945 | 0.0 |
| ... | ... | ... | ... |
| 5458 | 9990 | 999 | 0.0 |
| 5459 | 9992 | 918 | 0.0 |
| 5460 | 9995 | 940 | 0.0 |
| 5461 | 9996 | 992 | 0.0 |
| 5462 | 9998 | 963 | 0.0 |
5463 rows × 3 columns
| user_id | spend | treatment | |
|---|---|---|---|
| 0 | 1 | 926 | 0.0 |
| 1 | 2 | 922 | 0.0 |
| 2 | 3 | 1010 | 1.0 |
| 3 | 4 | 958 | 0.0 |
| 4 | 9 | 945 | 0.0 |
| ... | ... | ... | ... |
| 5458 | 9990 | 999 | 0.0 |
| 5459 | 9992 | 918 | 0.0 |
| 5460 | 9995 | 940 | 0.0 |
| 5461 | 9996 | 992 | 0.0 |
| 5462 | 9998 | 963 | 0.0 |
5463 rows × 3 columns
# total sales under three months of treatment for the beneficiary clients on June
# months under treatment (June, July and August)
base_6_3
| user_id | spend | treatment | |
|---|---|---|---|
| 0 | 1 | 1296 | 0.0 |
| 1 | 2 | 1296 | 0.0 |
| 2 | 4 | 1285 | 0.0 |
| 3 | 6 | 1500 | 1.0 |
| 4 | 9 | 1311 | 0.0 |
| ... | ... | ... | ... |
| 5472 | 9993 | 1589 | 1.0 |
| 5473 | 9995 | 1241 | 0.0 |
| 5474 | 9996 | 1308 | 0.0 |
| 5475 | 9997 | 1504 | 1.0 |
| 5476 | 9998 | 1253 | 0.0 |
5477 rows × 3 columns
| user_id | spend | treatment | |
|---|---|---|---|
| 0 | 1 | 1296 | 0.0 |
| 1 | 2 | 1296 | 0.0 |
| 2 | 4 | 1285 | 0.0 |
| 3 | 6 | 1500 | 1.0 |
| 4 | 9 | 1311 | 0.0 |
| ... | ... | ... | ... |
| 5472 | 9993 | 1589 | 1.0 |
| 5473 | 9995 | 1241 | 0.0 |
| 5474 | 9996 | 1308 | 0.0 |
| 5475 | 9997 | 1504 | 1.0 |
| 5476 | 9998 | 1253 | 0.0 |
5477 rows × 3 columns
# Kolmorogov - Smirnov Test: independent sample from common population distribution f(X)
ks_2samp(base_2_2['spend'][base_2_2.treatment == 1], base_2_2['spend'][base_2_2.treatment == 0])
# H0: samples come from same population distribution
# Pvalues is less than 5%, therefore the null hypothesis that both samples come from the same distribution is rejected.
KstestResult(statistic=0.8733908806150343, pvalue=8.7e-322, statistic_location=997, statistic_sign=-1)
KstestResult(statistic=0.8733908806150343, pvalue=8.7e-322, statistic_location=997, statistic_sign=-1)
test_1 = base_2_2.groupby("treatment")['spend'].agg(["count","mean","std","median","min","max"])
test_1
# Mean are different in both group
| count | mean | std | median | min | max | |
|---|---|---|---|---|---|---|
| treatment | ||||||
| 0.0 | 5006 | 949.386536 | 32.057687 | 949.0 | 819 | 1058 |
| 1.0 | 457 | 1047.463895 | 32.159405 | 1047.0 | 961 | 1150 |
| count | mean | std | median | min | max | |
|---|---|---|---|---|---|---|
| treatment | ||||||
| 0.0 | 5006 | 949.386536 | 32.057687 | 949.0 | 819 | 1058 |
| 1.0 | 457 | 1047.463895 | 32.159405 | 1047.0 | 961 | 1150 |
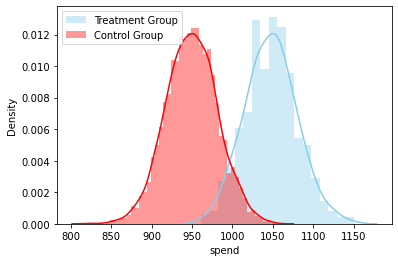
sns.distplot( base_2_2['spend'][base_2_2.treatment == 1] , color="skyblue", label="Treatment Group")
sns.distplot( base_2_2['spend'][base_2_2.treatment == 0] , color="red", label="Control Group")
plt.legend()
plt.show()
# Different densities corroborated the first test
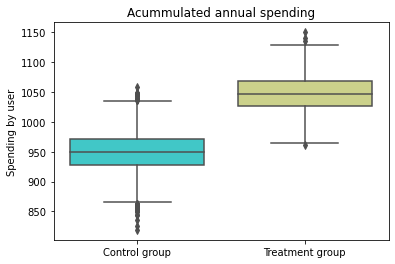
box = sns.boxplot(x="treatment", y="spend", data = base_2_2, palette='rainbow')
plt.ylabel('Spending by user')
plt.xlabel('')
plt.title('Acummulated annual spending')
(box.set_xticklabels(["Control group", "Treatment group"]));
jb1 = stats.jarque_bera(base_2_2["spend"][base_2_2.treatment == 1])
jb2 = stats.jarque_bera(base_2_2["spend"][base_2_2.treatment == 0])
print(jb1)
print(jb2)
# H0: samples come from Normal distribution
# Pvalues is larger than 5%, even 10%,, therefore, the null hypothesis that the
# samples were drawn from a normal distribution is not rejected.
SignificanceResult(statistic=1.0913765748896174, pvalue=0.5794428228359012)
SignificanceResult(statistic=0.9621851872679125, pvalue=0.6181076822166733)
t_value, t_test_p = stats.ttest_ind(base_2_2["spend"][base_2_2.treatment == 1],
base_2_2["spend"][base_2_2.treatment == 0], equal_var=False)
print('t_value = %.3f, t_test_p = %.3f' % (t_value, t_test_p),"\n")
# H0: samples come from same population distribution
# Non-parametric test that corroborates that both samples come from different distributions because p-vales is less 5%.
t_value = 62.426, t_test_p = 0.000
t_value, t_test_p = stats.mannwhitneyu(base_2_2["spend"][base_2_2.treatment == 1], base_2_2["spend"][base_2_2.treatment == 0])
print('statistics-t = %.3f, P_value= %.3f' % (t_value, t_test_p),"\n")
# H0: mean(y)treatment = mean(y)control
# p-vales is less 5%, hence H0 is rejected. The average sales of both groups is statistically different.
statistics-t = 2254562.000, P_value= 0.000
mod = sm.ols('spend ~ treatment', data=base_2_2)
res = mod.fit()
print(res.summary())
# For the beneficiaries of the month of February, the first two months undrer treatment increased sales by 98.07 dollars compared to the non-beneficiary group.
# This result is statistically significant at the 95% confidence level.
# the coefficient does not present important bias since the treatment and control groups are statistically comparable.
# Hence, no bias due to omission of relevant variables
OLS Regression Results
==============================================================================
Dep. Variable: spend R-squared: 0.418
Model: OLS Adj. R-squared: 0.418
Method: Least Squares F-statistic: 3918.
Date: Fri, 07 Apr 2023 Prob (F-statistic): 0.00
Time: 22:25:25 Log-Likelihood: -26695.
No. Observations: 5463 AIC: 5.339e+04
Df Residuals: 5461 BIC: 5.341e+04
Df Model: 1
Covariance Type: nonrobust
==============================================================================
coef std err t P>|t| [0.025 0.975]
------------------------------------------------------------------------------
Intercept 949.3865 0.453 2094.793 0.000 948.498 950.275
treatment 98.0774 1.567 62.591 0.000 95.005 101.149
==============================================================================
Omnibus: 0.827 Durbin-Watson: 1.984
Prob(Omnibus): 0.661 Jarque-Bera (JB): 0.780
Skew: 0.001 Prob(JB): 0.677
Kurtosis: 3.059 Cond. No. 3.64
==============================================================================
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
def abtest(base):
print(colored("Kolmorogov - Smirnov Test","cyan", attrs=["bold"]),"\n")
t_value, t_test_p = ks_2samp(base['spend'][base.treatment == 1], base['spend'][base.treatment == 0])
print('t_value = %.3f, t_test_p = %.3f' % (t_value, t_test_p),"\n")
print(colored("Summary Statistics","cyan", attrs=["bold"]),"\n")
test_1 = base.groupby("treatment")['spend'].agg(["count","mean","std","median","min","max"])
print(test_1, "\n")
print(colored("Jarque Bera Normality Test","cyan", attrs=["bold"]),"\n")
jb1 = stats.jarque_bera(base["spend"][base.treatment == 1])
jb2 = stats.jarque_bera(base["spend"][base.treatment == 0])
print(f'Jarque Bera (treatment group): {jb1}', "\n")
print(f'Jarque Bera (control group): {jb2}', "\n")
print(colored("Mann Whitney U Non parametric - Test","cyan", attrs=["bold"]),"\n")
t_value, t_test_p = stats.mannwhitneyu(base["spend"][base.treatment == 1], base["spend"][base.treatment == 0])
print('statistics-t = %.3f, P_value= %.3f' % (t_value, t_test_p),"\n")
print(colored("T-test mean","cyan", attrs=["bold"]),"\n")
t_value, t_test_p = stats.ttest_ind(base["spend"][base.treatment == 1],
base["spend"][base.treatment == 0], equal_var=False)
print('t_value = %.3f, t_test_p = %.3f' % (t_value, t_test_p),"\n")
print(colored("Densities Plot analysis", "cyan", attrs=["bold"]),"\n")
sns.distplot( base['spend'][base.treatment == 1] , color="darkblue", label="Treatment Group")
sns.distplot( base['spend'][base.treatment == 0] , color="red", label="Control Group")
plt.legend()
plt.show()
print(colored("Regression analysis", "cyan", attrs=["bold"]),"\n")
mod = sm.ols('spend ~ treatment', data=base)
res = mod.fit()
print(res.summary(),"\n")
print(colored("Pox Plot analysis", "cyan", attrs=["bold"]),"\n")
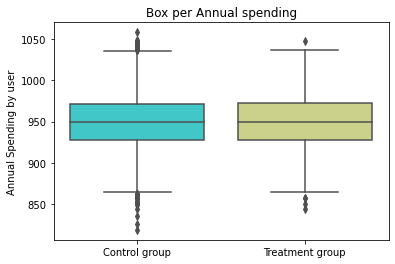
box = sns.boxplot(x="treatment", y="spend", data= base, palette='rainbow')
plt.ylabel('Annual Spending by user')
plt.xlabel('')
plt.title('Box per Annual spending')
(box.set_xticklabels(["Control group", "Treatment group"]))
abtest(base_2_2)
Kolmorogov - Smirnov Test
t_value = 0.873, t_test_p = 0.000
Summary Statistics
count mean std median min max
treatment
0.0 5006 949.386536 32.057687 949.0 819 1058
1.0 457 1047.463895 32.159405 1047.0 961 1150
Jarque Bera Normality Test
Jarque Bera (treatment group): SignificanceResult(statistic=1.0913765748896174, pvalue=0.5794428228359012)
Jarque Bera (control group): SignificanceResult(statistic=0.9621851872679125, pvalue=0.6181076822166733)
Mann Whitney U Non parametric - Test
statistics-t = 2254562.000, P_value= 0.000
T-test mean
t_value = 62.426, t_test_p = 0.000
Densities Plot analysis
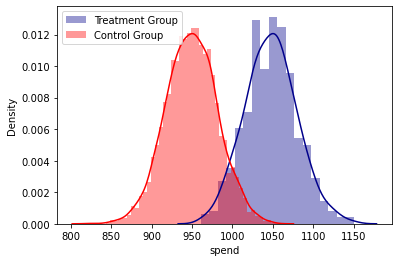
Regression analysis
OLS Regression Results
==============================================================================
Dep. Variable: spend R-squared: 0.418
Model: OLS Adj. R-squared: 0.418
Method: Least Squares F-statistic: 3918.
Date: Fri, 07 Apr 2023 Prob (F-statistic): 0.00
Time: 22:25:30 Log-Likelihood: -26695.
No. Observations: 5463 AIC: 5.339e+04
Df Residuals: 5461 BIC: 5.341e+04
Df Model: 1
Covariance Type: nonrobust
==============================================================================
coef std err t P>|t| [0.025 0.975]
------------------------------------------------------------------------------
Intercept 949.3865 0.453 2094.793 0.000 948.498 950.275
treatment 98.0774 1.567 62.591 0.000 95.005 101.149
==============================================================================
Omnibus: 0.827 Durbin-Watson: 1.984
Prob(Omnibus): 0.661 Jarque-Bera (JB): 0.780
Skew: 0.001 Prob(JB): 0.677
Kurtosis: 3.059 Cond. No. 3.64
==============================================================================
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
Pox Plot analysis
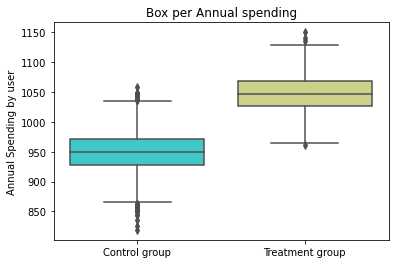
#Positive causal effect of the marketing strategy over time
coef = pd.DataFrame(columns=('months','causal_effect', 'lower','upper'))
for j in range(2,12):
mod = sm.ols('spend ~ treatment', data=globals()[f'base_2_{j}'])
res = mod.fit()
coef.loc[j-2,'lower']=res.conf_int(alpha=0.05, cols=None)[0][1]
coef.loc[j-2,'upper']=res.conf_int(alpha=0.05, cols=None)[1][1]
coef.loc[j-2,'causal_effect']=res.params[1]
coef.loc[j-2,'months']= j
coef
| months | causal_effect | lower | upper | |
|---|---|---|---|---|
| 0 | 2 | 98.077359 | 95.005482 | 101.149236 |
| 1 | 3 | 198.97023 | 195.249432 | 202.691028 |
| 2 | 4 | 299.287842 | 295.014514 | 303.56117 |
| 3 | 5 | 400.08003 | 395.333792 | 404.826269 |
| 4 | 6 | 500.627491 | 495.414822 | 505.840161 |
| 5 | 7 | 602.782273 | 597.151287 | 608.413259 |
| 6 | 8 | 703.938904 | 697.942304 | 709.935505 |
| 7 | 9 | 805.135222 | 798.744774 | 811.525669 |
| 8 | 10 | 1005.810219 | 998.792173 | 1012.828265 |
| 9 | 11 | 1005.810219 | 998.792173 | 1012.828265 |
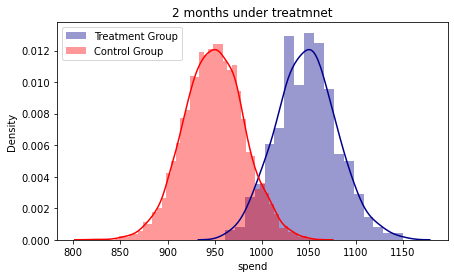
fig, axes = plt.subplots(figsize=(7, 4))
ax1 = sns.distplot( base_2_2['spend'][base_2_2.treatment == 1] , color="darkblue", label="Treatment Group")
ax1 = sns.distplot( base_2_2['spend'][base_2_2.treatment == 0] , color="red", label="Control Group")
plt.title('2 months under treatmnet')
plt.legend()
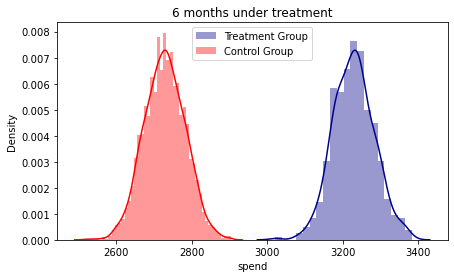
fig, ax = plt.subplots(figsize=(7, 4))
ax2 = sns.distplot( base_2_6['spend'][base_2_6.treatment == 1] , color="darkblue", label="Treatment Group")
ax2 = sns.distplot( base_2_6['spend'][base_2_6.treatment == 0] , color="red", label="Control Group")
plt.title('6 months under treatment')
plt.legend()
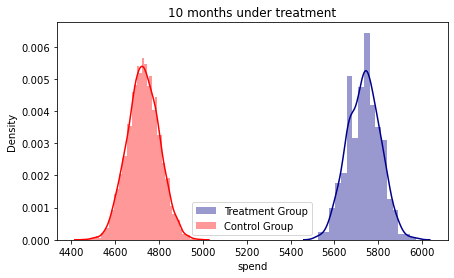
fig, ax = plt.subplots(figsize=(7, 4))
ax3 = sns.distplot( base_2_10['spend'][base_2_10.treatment == 1] , color="darkblue", label="Treatment Group")
ax3 = sns.distplot( base_2_10['spend'][base_2_10.treatment == 0] , color="red", label="Control Group")
plt.title('10 months under treatment')
plt.legend()
#The densities show an increasing gap according to the months accumulated under the treatment.
#Treatment clients on february
<matplotlib.legend.Legend at 0x1f0445ac8e0>
base_2_10 = causal_signup_month(base,2,10)
# AB test for ten months under treatment for the beneficiary clients on February
abtest(base_2_10)
Kolmorogov - Smirnov Test
t_value = 1.000, t_test_p = 0.000
Summary Statistics
count mean std median min max
treatment
0.0 5006 4728.382341 73.222369 4728.0 4456 4988
1.0 457 5734.192560 73.657384 5737.0 5525 5970
Jarque Bera Normality Test
Jarque Bera (treatment group): SignificanceResult(statistic=0.4653152162176708, pvalue=0.7924248469227017)
Jarque Bera (control group): SignificanceResult(statistic=0.6594857909581255, pvalue=0.7191085957374495)
Mann Whitney U Non parametric - Test
statistics-t = 2287742.000, P_value= 0.000
T-test mean
t_value = 279.577, t_test_p = 0.000
Densities Plot analysis
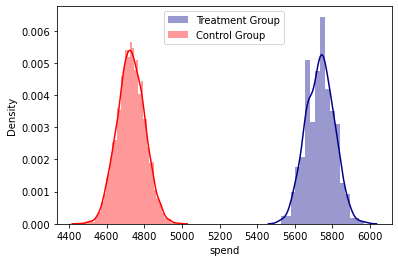
Regression analysis
OLS Regression Results
==============================================================================
Dep. Variable: spend R-squared: 0.935
Model: OLS Adj. R-squared: 0.935
Method: Least Squares F-statistic: 7.894e+04
Date: Fri, 07 Apr 2023 Prob (F-statistic): 0.00
Time: 22:25:53 Log-Likelihood: -31209.
No. Observations: 5463 AIC: 6.242e+04
Df Residuals: 5461 BIC: 6.243e+04
Df Model: 1
Covariance Type: nonrobust
==============================================================================
coef std err t P>|t| [0.025 0.975]
------------------------------------------------------------------------------
Intercept 4728.3823 1.035 4566.656 0.000 4726.353 4730.412
treatment 1005.8102 3.580 280.960 0.000 998.792 1012.828
==============================================================================
Omnibus: 0.918 Durbin-Watson: 2.000
Prob(Omnibus): 0.632 Jarque-Bera (JB): 0.947
Skew: 0.003 Prob(JB): 0.623
Kurtosis: 2.936 Cond. No. 3.64
==============================================================================
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
Pox Plot analysis
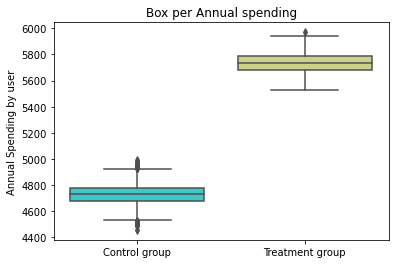
#Placebo Test: Treatment groups are artificially created from control group.
# Beneficiary units are randomly selected from the control group
# artificial beneficiary clients of february
j = 2
pbo_base0 = base[base.treatment == 0].drop('treatment', axis =1)
pbo_base1 = pbo_base0.drop_duplicates(subset=['user_id'])
pbo_base1 = pbo_base1.sample(frac=0.25, replace=False, random_state=100)
pbo_base1 = pbo_base1[['user_id']]
pbo_base1['treatment'] = 1
placebo_base = pd.merge(pbo_base0, pbo_base1, how="left", on=["user_id"])
placebo_base['treatment'] = placebo_base['treatment'].fillna(0)
placebo_base.loc[placebo_base.treatment == 1, 'signup_month'] = j
globals()[f'placebo_base_{j}'] = placebo_base
# first two months under treatment for february artifical benficiaries
placebo_2_2 = causal_signup_month(placebo_base_2,2,1)
placebo_base_2.describe()
| user_id | signup_month | month | spend | treatment | |
|---|---|---|---|---|---|
| count | 60072.000000 | 60072.000000 | 60072.000000 | 60072.000000 | 60072.000000 |
| mean | 4972.272473 | 0.500200 | 6.500000 | 434.853443 | 0.250100 |
| std | 2890.220110 | 0.866148 | 3.452081 | 41.062485 | 0.433074 |
| min | 1.000000 | 0.000000 | 1.000000 | 286.000000 | 0.000000 |
| 25% | 2448.000000 | 0.000000 | 3.750000 | 404.000000 | 0.000000 |
| 50% | 5010.000000 | 0.000000 | 6.500000 | 435.000000 | 0.000000 |
| 75% | 7475.000000 | 2.000000 | 9.250000 | 466.000000 | 1.000000 |
| max | 9998.000000 | 2.000000 | 12.000000 | 566.000000 | 1.000000 |
#Placebo test for artificial beneficiary group on february
abtest(placebo_2_2)
#It can be seen that both groups are statistically equal.
# There is no evidence that the artificial treatment group has increased their purchases.
Kolmorogov - Smirnov Test
t_value = 0.019, t_test_p = 0.867
Summary Statistics
count mean std median min max
treatment
0.0 3754 949.257326 32.146578 949.5 819 1058
1.0 1252 949.773962 31.799294 949.0 844 1048
Jarque Bera Normality Test
Jarque Bera (treatment group): SignificanceResult(statistic=0.8424322407805283, pvalue=0.6562482574525269)
Jarque Bera (control group): SignificanceResult(statistic=2.533146164438528, pvalue=0.2817956595520864)
Mann Whitney U Non parametric - Test
statistics-t = 2371435.000, P_value= 0.628
T-test mean
t_value = 0.496, t_test_p = 0.620
Densities Plot analysis
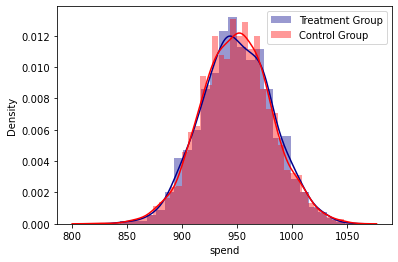
Regression analysis
OLS Regression Results
==============================================================================
Dep. Variable: spend R-squared: 0.000
Model: OLS Adj. R-squared: -0.000
Method: Least Squares F-statistic: 0.2438
Date: Fri, 07 Apr 2023 Prob (F-statistic): 0.621
Time: 22:26:01 Log-Likelihood: -24461.
No. Observations: 5006 AIC: 4.893e+04
Df Residuals: 5004 BIC: 4.894e+04
Df Model: 1
Covariance Type: nonrobust
==============================================================================
coef std err t P>|t| [0.025 0.975]
------------------------------------------------------------------------------
Intercept 949.2573 0.523 1814.120 0.000 948.232 950.283
treatment 0.5166 1.046 0.494 0.621 -1.535 2.568
==============================================================================
Omnibus: 1.004 Durbin-Watson: 1.996
Prob(Omnibus): 0.605 Jarque-Bera (JB): 0.957
Skew: -0.010 Prob(JB): 0.620
Kurtosis: 3.065 Cond. No. 2.48
==============================================================================
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
Pox Plot analysis